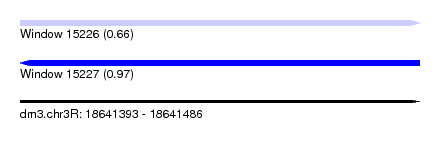
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,641,393 – 18,641,486 |
| Length | 93 |
| Max. P | 0.972444 |

| Location | 18,641,393 – 18,641,486 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.91 |
| Shannon entropy | 0.35017 |
| G+C content | 0.56235 |
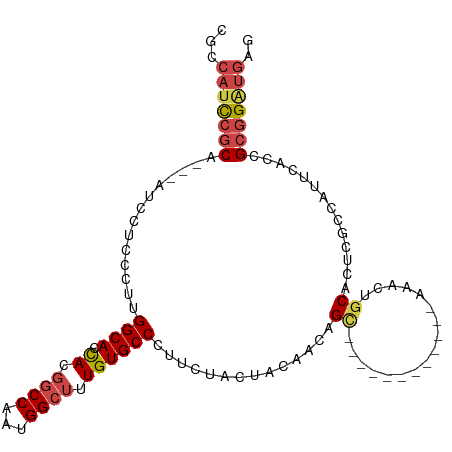
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
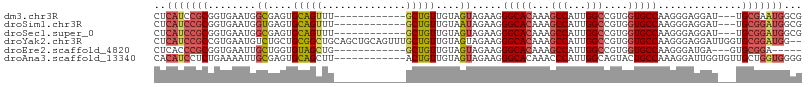
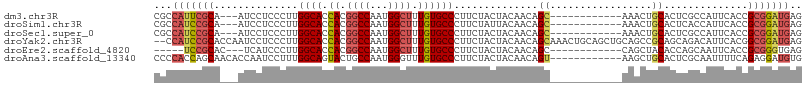
>dm3.chr3R 18641393 93 + 27905053 CUCAUCCGCGGUGAAUGGCGAGUGCAGUUU------------GCUGUUGUAGUAGAAGGGCACAAAGCCAUUGGCCGUGGUGCCAAGGGAGGAU---UGCGAAUGGCG .....(((((((.((((((..((((..(((------------((((...)))))))...))))...)))))).))))))).((((.(.((...)---).)...)))). ( -39.60, z-score = -3.35, R) >droSim1.chr3R 18453265 93 + 27517382 CUCAUCCGCGGUGAAUGGUGAGUGCAGUUU------------GCUGUUGUAAUAGAAGGGCACAAAGCCAUUGGCCGUGGUGCCAAGGGAGGAU---UGCGGAUGGCG .(((((((((((.((((((..((((.....------------.((((....))))....))))...)))))).)))......((......))..---.)))))))).. ( -35.90, z-score = -2.34, R) >droSec1.super_0 19002121 93 + 21120651 CUCAUCCGCGGUGAAUGGCGAGUGCAGUUU------------GCUGUUGUAGUAGAAGGGCACAAAGCCAUUGGCCGUGGUGCCAAGGGAGGAU---UGCGGAUGGCG .(((((((((((.((((((..((((..(((------------((((...)))))))...))))...)))))).)))......((......))..---.)))))))).. ( -41.10, z-score = -3.67, R) >droYak2.chr3R 19494224 106 + 28832112 CUCAUCCGCCGUGAAUGUCUGCUGCGGCUGCAGCUGCAGUUUGCUGUUGUAGUAGAAGGGCACAAAGCCAUUGGCCGUGGUGCCAAGGGAGGAUUGGUGCGGAUGG-- ..(((((((((((....(((((((((((.(((((....).)))).)))))))))))....)))....((.(((((......))))).)).......).))))))).-- ( -45.30, z-score = -1.88, R) >droEre2.scaffold_4820 1039359 88 - 10470090 CUCACCCGCGGUGAAUUGCUGGUGUAGCUG------------GCUGUUGUAGUAGAAGGGCACAAAGCCAUUGGCCGUGGUGCCAAGGGAUGA---GUGCGGA----- ((((((((((((.(.((((....)))).).------------)))))...........(((((...(((...)))....)))))..))).)))---)......----- ( -29.20, z-score = 0.22, R) >droAna3.scaffold_13340 11022829 96 - 23697760 CACAUCCUCUGAAAAUUGCGAGUGCAGCUU------------ACUGUUGUAGUAGAAGGGCACAAACCCAUUGGCAGUACUGCCAAAGGAUUGGUGUUGCUGGUGGGG .....................((((..(((------------.(((......)))))).))))...(((((..((((((((.((...))...))))))))..))))). ( -29.30, z-score = -0.27, R) >consensus CUCAUCCGCGGUGAAUGGCGAGUGCAGCUU____________GCUGUUGUAGUAGAAGGGCACAAAGCCAUUGGCCGUGGUGCCAAGGGAGGAU___UGCGGAUGGCG ..(((((((........((....(((((..............)))))....)).....(((((...(((...)))....)))))..............)))))))... (-21.40 = -22.24 + 0.84)
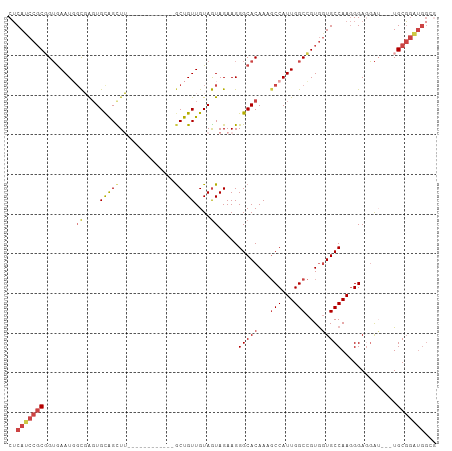
| Location | 18,641,393 – 18,641,486 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.91 |
| Shannon entropy | 0.35017 |
| G+C content | 0.56235 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
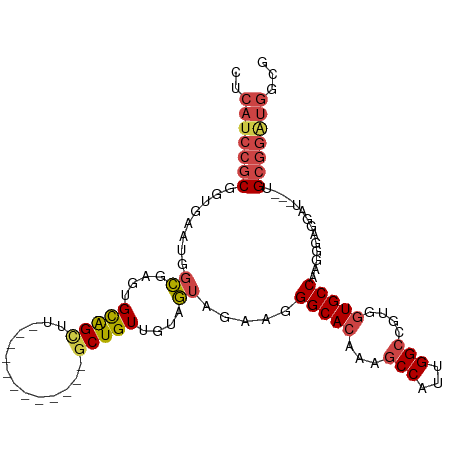
>dm3.chr3R 18641393 93 - 27905053 CGCCAUUCGCA---AUCCUCCCUUGGCACCACGGCCAAUGGCUUUGUGCCCUUCUACUACAACAGC------------AAACUGCACUCGCCAUUCACCGCGGAUGAG ...(((((((.---..........(((......)))((((((...((((.......((.....)).------------.....))))..))))))....))))))).. ( -25.32, z-score = -2.19, R) >droSim1.chr3R 18453265 93 - 27517382 CGCCAUCCGCA---AUCCUCCCUUGGCACCACGGCCAAUGGCUUUGUGCCCUUCUAUUACAACAGC------------AAACUGCACUCACCAUUCACCGCGGAUGAG ...(((((((.---..........(((((...((((...))))..)))))..............((------------.....))..............))))))).. ( -26.90, z-score = -3.24, R) >droSec1.super_0 19002121 93 - 21120651 CGCCAUCCGCA---AUCCUCCCUUGGCACCACGGCCAAUGGCUUUGUGCCCUUCUACUACAACAGC------------AAACUGCACUCGCCAUUCACCGCGGAUGAG ...(((((((.---..........(((......)))((((((...((((.......((.....)).------------.....))))..))))))....))))))).. ( -28.22, z-score = -3.26, R) >droYak2.chr3R 19494224 106 - 28832112 --CCAUCCGCACCAAUCCUCCCUUGGCACCACGGCCAAUGGCUUUGUGCCCUUCUACUACAACAGCAAACUGCAGCUGCAGCCGCAGCAGACAUUCACGGCGGAUGAG --.(((((((..............(((((...((((...))))..)))))..(((.........((.....)).(((((....))))))))........))))))).. ( -36.00, z-score = -2.67, R) >droEre2.scaffold_4820 1039359 88 + 10470090 -----UCCGCAC---UCAUCCCUUGGCACCACGGCCAAUGGCUUUGUGCCCUUCUACUACAACAGC------------CAGCUACACCAGCAAUUCACCGCGGGUGAG -----......(---((((((.(((((......)))))((((((((((.........))))).)))------------))(((.....)))..........))))))) ( -26.00, z-score = -1.99, R) >droAna3.scaffold_13340 11022829 96 + 23697760 CCCCACCAGCAACACCAAUCCUUUGGCAGUACUGCCAAUGGGUUUGUGCCCUUCUACUACAACAGU------------AAGCUGCACUCGCAAUUUUCAGAGGAUGUG ...(((((((..(((.(((((.((((((....)))))).))))).)))......((((.....)))------------).))))..(((..........)))...))) ( -26.60, z-score = -1.49, R) >consensus CGCCAUCCGCA___AUCCUCCCUUGGCACCACGGCCAAUGGCUUUGUGCCCUUCUACUACAACAGC____________AAACUGCACUCGCCAUUCACCGCGGAUGAG ...(((((((............(((((......))))).(((.....))).................................................))))))).. (-18.00 = -18.75 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:04 2011