| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,621,294 – 18,621,384 |
| Length | 90 |
| Max. P | 0.597976 |

| Location | 18,621,294 – 18,621,384 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.40297 |
| G+C content | 0.34101 |
| Mean single sequence MFE | -14.21 |
| Consensus MFE | -9.90 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
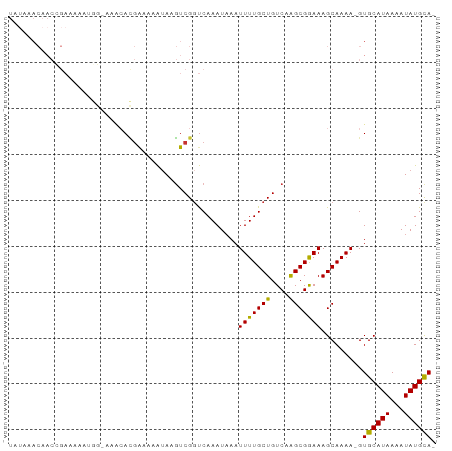
>dm3.chr3R 18621294 90 + 27905053 UAUAAACAACCGAAAAAUGG-GAACACGAAAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAAGCGGAAAGCAAAA-GUGCAUAAAAUAUGCA- ........(((((....((.-...))...........)))))........(((((((.((.....)).)))))))-.((((((...))))))- ( -15.26, z-score = -1.42, R) >droSim1.chr3R 18433240 90 + 27517382 UAUAAACAACCGAAAAAUGG-GAGCACGAAAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAAGCGGAAAGCAAAA-GUGCAUAAAAUAUGCA- .........((........)-).((((.........(.(((.((((.....))))))).)..((.....))....-))))............- ( -14.80, z-score = -0.84, R) >droSec1.super_0 18982026 92 + 21120651 UAUAAACAACCGAAAAAUGGGCACCACGAAAAAUAAGUCGUUCAAAUUAAUUUUGCUGUCAAGCGGAAAGCAAAAAGUGCAUAAAAUAUGCA- .........(((.....)))((((.((((........)))).........(((((((.((.....)).))))))).))))............- ( -15.50, z-score = -0.84, R) >droYak2.chr3R 19472794 90 + 28832112 UAUAAACAACUGAAAAAUGG-GAACACGAAAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAAGCGGAAAGCAAAA-GUGCAUAAAAUAUGUA- ..........(((....((.-...))(((........))).)))......(((((((.((.....)).)))))))-.((((((...))))))- ( -11.70, z-score = -0.38, R) >droEre2.scaffold_4820 1018134 89 - 10470090 UAUAAACAACUGAAAA-UGGGAAC-ACGAAAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAAGCGGAAAGCAAAA-GUGCAUAAAAUAUGCA- ..........(((...-((....)-)(((........))).)))......(((((((.((.....)).)))))))-.((((((...))))))- ( -13.70, z-score = -0.91, R) >droAna3.scaffold_13340 11002849 78 - 23697760 -------------AGGCCCCCAAAAAGAACACGAAACUCGGCCAAAUAAAUUUUGCUGUCAAGCGAAAAGCAAAA-GUGCAUAAAAUAUGCA- -------------.((((.....................)))).......(((((((....))))))).......-.((((((...))))))- ( -15.40, z-score = -1.38, R) >dp4.chr2 24592046 70 - 30794189 ----------------------AACAUGGCAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAGGCGAAAAGCAAAA-GUGCAUAAAAUAUGCAC ----------------------....((((......))))..........(((((((.((....))..)))))))-(((((((...))))))) ( -14.30, z-score = -1.67, R) >droPer1.super_0 11794097 87 - 11822988 UAAGAACAACAGAAAA-----AAACAUGGUAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAGGCGAAAAGCAAAA-GUGCAUAAAAUAUGCAC ................-----...............(.(((.((((.....))))))).)..((.....))....-(((((((...))))))) ( -13.00, z-score = -1.32, R) >consensus UAUAAACAACCGAAAAAUGG_AAACACGAAAAAUAAGUCGGUCAAAUAAAUUUUGCUGUCAAGCGGAAAGCAAAA_GUGCAUAAAAUAUGCA_ ..................................................(((((((....))))))).........((((((...)))))). ( -9.90 = -9.38 + -0.53)


| Location | 18,621,294 – 18,621,384 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.15 |
| Shannon entropy | 0.40297 |
| G+C content | 0.34101 |
| Mean single sequence MFE | -13.85 |
| Consensus MFE | -8.88 |
| Energy contribution | -8.78 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
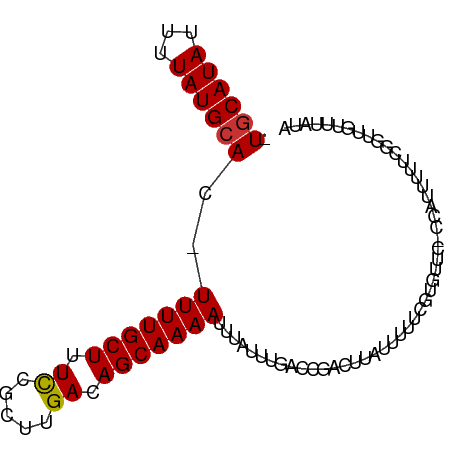
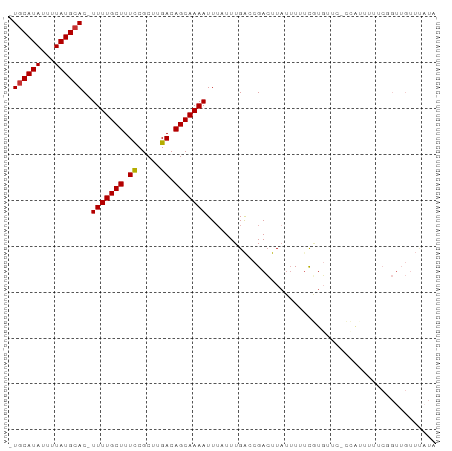
>dm3.chr3R 18621294 90 - 27905053 -UGCAUAUUUUAUGCAC-UUUUGCUUUCCGCUUGACAGCAAAAUUUAUUUGACCGACUUAUUUUUCGUGUUC-CCAUUUUUCGGUUGUUUAUA -((((((...)))))).-(((((((.((.....)).)))))))..((..(((((((................-.......)))))))..)).. ( -15.30, z-score = -2.04, R) >droSim1.chr3R 18433240 90 - 27517382 -UGCAUAUUUUAUGCAC-UUUUGCUUUCCGCUUGACAGCAAAAUUUAUUUGACCGACUUAUUUUUCGUGCUC-CCAUUUUUCGGUUGUUUAUA -((((((...)))))).-(((((((.((.....)).)))))))..((..(((((((................-.......)))))))..)).. ( -15.30, z-score = -1.89, R) >droSec1.super_0 18982026 92 - 21120651 -UGCAUAUUUUAUGCACUUUUUGCUUUCCGCUUGACAGCAAAAUUAAUUUGAACGACUUAUUUUUCGUGGUGCCCAUUUUUCGGUUGUUUAUA -((((((...))))))..(((((((.((.....)).)))))))......(((((((((........((((...)))).....))))))))).. ( -18.42, z-score = -1.81, R) >droYak2.chr3R 19472794 90 - 28832112 -UACAUAUUUUAUGCAC-UUUUGCUUUCCGCUUGACAGCAAAAUUUAUUUGACCGACUUAUUUUUCGUGUUC-CCAUUUUUCAGUUGUUUAUA -............((((-(((((((.((.....)).)))))))...........((........))))))..-.................... ( -8.90, z-score = -0.38, R) >droEre2.scaffold_4820 1018134 89 + 10470090 -UGCAUAUUUUAUGCAC-UUUUGCUUUCCGCUUGACAGCAAAAUUUAUUUGACCGACUUAUUUUUCGU-GUUCCCA-UUUUCAGUUGUUUAUA -((((((...)))))).-(((((((.((.....)).))))))).......((((((........))).-)))....-................ ( -12.40, z-score = -1.26, R) >droAna3.scaffold_13340 11002849 78 + 23697760 -UGCAUAUUUUAUGCAC-UUUUGCUUUUCGCUUGACAGCAAAAUUUAUUUGGCCGAGUUUCGUGUUCUUUUUGGGGGCCU------------- -((((((...)))))).-(((((((..((....)).))))))).......(((((.....)....(((....))))))).------------- ( -14.60, z-score = 0.46, R) >dp4.chr2 24592046 70 + 30794189 GUGCAUAUUUUAUGCAC-UUUUGCUUUUCGCCUGACAGCAAAAUUUAUUUGACCGACUUAUUUGCCAUGUU---------------------- (((((((...)))))))-(((((((..((....)).)))))))............................---------------------- ( -11.90, z-score = -1.38, R) >droPer1.super_0 11794097 87 + 11822988 GUGCAUAUUUUAUGCAC-UUUUGCUUUUCGCCUGACAGCAAAAUUUAUUUGACCGACUUAUUUACCAUGUUU-----UUUUCUGUUGUUCUUA (((((((...)))))))-(((((((..((....)).))))))).......((.((((...............-----......)))).))... ( -14.00, z-score = -2.29, R) >consensus _UGCAUAUUUUAUGCAC_UUUUGCUUUCCGCUUGACAGCAAAAUUUAUUUGACCGACUUAUUUUUCGUGUUC_CCAUUUUUCGGUUGUUUAUA .((((((...))))))..(((((((.((.....)).))))))).................................................. ( -8.88 = -8.78 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:03 2011