| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,609,520 – 18,609,612 |
| Length | 92 |
| Max. P | 0.945869 |

| Location | 18,609,520 – 18,609,612 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 71.48 |
| Shannon entropy | 0.53949 |
| G+C content | 0.40292 |
| Mean single sequence MFE | -21.64 |
| Consensus MFE | -13.41 |
| Energy contribution | -14.93 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.945869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
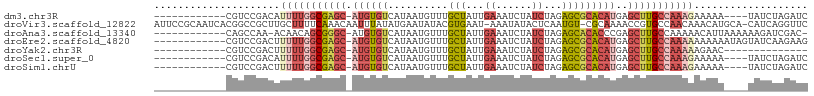
>dm3.chr3R 18609520 92 - 27905053 ------------CGUCCGACAUUUUGGCGAGC-AUGUGUCAUAAUGUUUGCUAUUGAAAUCUAUCUAGAGCGCACAUGAGCUUGCCAAAGAAAAA----UAUCUAGAUC ------------.........(((((((((((-((((((..........(((...((......))...)))))))))..))))))))))).....----.......... ( -23.20, z-score = -1.51, R) >droVir3.scaffold_12822 3694626 106 + 4096053 AUUCCGCAAUCACGGCCGCUUGCUUUUCAAACAAUUUAUAUGAAUAUACGUGAAU-AAAUAUACUCAAUGU-CGCAAAACCGUGCCAACAAACAUGCA-CAUCAGGUUC ...((((..(((((((.....))..((((...........))))....)))))..-......((.....))-.))......((((..........)))-)....))... ( -12.60, z-score = 1.29, R) >droAna3.scaffold_13340 10989324 94 + 23697760 ------------CAGCCAA-ACAACAGCGGGC-AUGUGUCAUAAUGUUUGCUAUUGAAAUCUAUCUAGAGCACACCCGAGCUUGCCAAAAACAUUAAAAAAGAUCGAC- ------------..((...-.(((.(((((((-((........))))))))).)))...(((....))))).....(((.(((................))).)))..- ( -16.49, z-score = -0.22, R) >droEre2.scaffold_4820 1006232 96 + 10470090 ------------CGUCCGACUUUUUGGCGAGC-AUGUGUCAUAAUGUUUGCUAUUGAAAUCUAUCUAGAGCGCACAUGAGCUUGCCAAAAAAAAAAUAGUAUCAAGAAG ------------........((((((((((((-((((((..........(((...((......))...)))))))))..)))))))))))).................. ( -24.20, z-score = -2.01, R) >droYak2.chr3R 19460805 82 - 28832112 ------------CGUCCGACUUUUUGGCGAGC-AUGUGUCAUAAUGUUUGCUAUUGAAAUCUAUCUAGAGCGCACAUGAGCUUGCCAAAAAGAAC-------------- ------------.......(((((((((((((-((((((..........(((...((......))...)))))))))..)))))))))))))...-------------- ( -27.50, z-score = -3.70, R) >droSec1.super_0 18970350 92 - 21120651 ------------CGUCCGACAUUUUGGCGAGC-AUGUGUCAUAAUGUUUGCUAUUGAAAUCUAUCUAGAGCGCACAUGAGCUUGCCAAAGAAAAA----UAUCUAGAUC ------------.........(((((((((((-((((((..........(((...((......))...)))))))))..))))))))))).....----.......... ( -23.20, z-score = -1.51, R) >droSim1.chrU 10960857 92 - 15797150 ------------CGUCCGACUUUUUGGCGAGC-AUGUGUCAUAAUGUUUGCUAUUGAAAUCUAUCUAGAGCGCACAUGAGCUUGCCAAAGAAAAA----UAUCUAGAUC ------------........((((((((((((-((((((..........(((...((......))...)))))))))..))))))))))))....----.......... ( -24.30, z-score = -1.92, R) >consensus ____________CGUCCGACUUUUUGGCGAGC_AUGUGUCAUAAUGUUUGCUAUUGAAAUCUAUCUAGAGCGCACAUGAGCUUGCCAAAAAAAAA____UAUCUAGAUC .....................(((((((((((..(((((..........(((...((......))...))))))))...)))))))))))................... (-13.41 = -14.93 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:01 2011