
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,608,675 – 18,608,765 |
| Length | 90 |
| Max. P | 0.526796 |

| Location | 18,608,675 – 18,608,765 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 67.54 |
| Shannon entropy | 0.61932 |
| G+C content | 0.43063 |
| Mean single sequence MFE | -17.10 |
| Consensus MFE | -8.12 |
| Energy contribution | -8.02 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
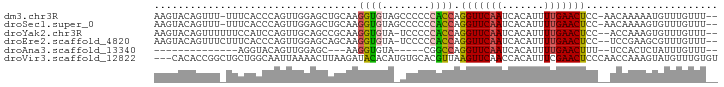
>dm3.chr3R 18608675 90 - 27905053 AAGUACAGUUU-UUUCACCCAGUUGGAGCUGCAAGGUGUAGCCCCCCACCAGGUUCAAUCACAUUUUGAACUCC-AACAAAAAUGUUUGUUU-- ...........-.........(((((((...((((((((((((........)))).....))))))))..))))-)))..............-- ( -20.30, z-score = -1.33, R) >droSec1.super_0 18969546 90 - 21120651 AAGUACAGUUU-UUUCACCCAGUUGGAGCUGCAAGGUGUAGCCCCCCACCAGGUUCAAUCACAUUUUGAACUCC-AACAAAAGUGUUUGUUU-- ...........-...(((...(((((((...((((((((((((........)))).....))))))))..))))-)))....))).......-- ( -20.70, z-score = -1.29, R) >droYak2.chr3R 19459905 89 - 28832112 AAGUACAGUUUUUUCCAUCCAGUUGCAGCCGCAAGGUGUA-UCCCCCACCAGGUUCAAUCACAUUUUGAACUCC--ACCAAAGUGUUUGUUU-- ....((((.............((((.((((....((((..-.....)))).)))))))).((((((((......--..))))))))))))..-- ( -16.20, z-score = -1.65, R) >droEre2.scaffold_4820 1005342 89 + 10470090 AAGUACAGUUUCUUUCACCCAGUUGGAGCAGCAAGGUGUA-UCCCCCACCAGGUUCAAUCACAUUUUGAACUCC--UCCGAAGCGUUUGUUU-- .......(((((...(.....)..((((......((((..-.....)))).(((((((.......))))))).)--))))))))........-- ( -17.70, z-score = -0.95, R) >droAna3.scaffold_13340 10988457 68 + 23697760 --------------AGGUACAGUUGGAGC---AAGGUGUA-----CGGCCAGGUUCAAUCACAUUUUGAACUUU--UCCACUCUAUUUGUUU-- --------------..(((((.((.....---.)).))))-----)((..((((((((.......)))))))).--.)).............-- ( -16.30, z-score = -1.42, R) >droVir3.scaffold_12822 3693884 91 + 4096053 ---CACACCGGCUGCUGGCAAUUAAAACUUAAGAUACACAUGUGCACGUUAAGUUCAACCACAUUUCGAACUCCCAACCAAAGUAUGUUUGUGU ---....((((...))))...............(((((.((((((..(((.(((((...........)))))...)))....)))))).))))) ( -11.40, z-score = 1.79, R) >consensus AAGUACAGUUU_UUUCACCCAGUUGGAGCUGCAAGGUGUA_CCCCCCACCAGGUUCAAUCACAUUUUGAACUCC__ACCAAAGUGUUUGUUU__ ..................................((((........)))).(((((((.......)))))))...................... ( -8.12 = -8.02 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:00 2011