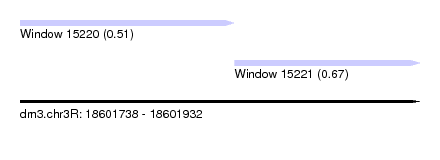
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,601,738 – 18,601,932 |
| Length | 194 |
| Max. P | 0.665071 |

| Location | 18,601,738 – 18,601,842 |
|---|---|
| Length | 104 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Shannon entropy | 0.45162 |
| G+C content | 0.52103 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -16.53 |
| Energy contribution | -15.55 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18601738 104 + 27905053 CCGCUGUGGUUAUCCACUGCGGUUCCGGACCAAAUCGUGGCCAUUAUAUAUCCAUUGUGAAGAGUCACGGACUGUGGCUACUAUUCGACGAUGAUAUGGUGGAC---------------- ((((.((((....)))).)))).(((((.(((.....)))))....((((((.((((((((.((((((.....))))))....))).)))))))))))..))).---------------- ( -37.60, z-score = -2.41, R) >anoGam1.chr3L 3957722 112 - 41284009 UGGCGGUGGUCAUACACUGCGGCUCGGGCCCCAACCGUGGCCAUUACAUUAGCAUUGUCAAAAGUCACGGUUUCUGGCUGUUGUUUGACGACGAUAUGGUUGAUGUAAGUCA-------- ..((((((......))))))(((((((.......))).)))).((((((((((..((((...(((((.......)))))((((.....))))))))..))))))))))....-------- ( -40.50, z-score = -1.72, R) >droGri2.scaffold_15074 3396617 104 + 7742996 CGGCUGUCGUUAUUCAUUGCGGUUCGGGUCCGAAUCGUGGCCAUUACAUCAGCAUUGUGAAGAGUCACGGACUCUGGCUACUGUUCGACGAUGACAUGGUCGAU---------------- (((((((.((((((....((((((((....))))))((((((.(((((.......)))))((((((...)))))))))))).)).....)))))))))))))..---------------- ( -38.80, z-score = -2.07, R) >droMoj3.scaffold_6540 1107820 104 - 34148556 CCGCAGUCGUCAUACACUGCGGCUCGGGCCCGAAUCGUGGCCAUUACAUAAGCAUUGUCAAGAGUCACGGGCUUUGGCUGCUCUUCGAUGACGACAUGGUUGAC---------------- (((..((((((((.....((((((..(((((.....(((((..(((((.......))).))..))))))))))..))))))......)))))))).))).....---------------- ( -40.50, z-score = -2.58, R) >droVir3.scaffold_12822 3683183 104 - 4096053 CCGCGGUGGUAAUUCACUGCGGAUCGGGACCGAAUCGCGGCCAUUACAUAAGCAUUGUGAAGAGUCACGGGCUCUGGCUGCUCUUCGACGACGACAUGGUCGAC---------------- (((((((((....)))))))))........((((..((((((.(((((.......)))))(((((.....)))))))))))..)))).((((......))))..---------------- ( -43.90, z-score = -3.24, R) >droWil1.scaffold_181089 10937061 104 - 12369635 CCGCCGUGGUCAUCCACUGUGGCUCUGGACCGAAUCGUGGCCAUUAUAUUAGCAUUGUCAAGAGCCACGGCCUGUGGUUGCUGUUCGACGAUGAUAUGGUCGAU---------------- .(((((((.(((((..((((((((((((.(((.....)))))...(((.......)))..))))))))))......((((.....)))))))))))))).))..---------------- ( -33.40, z-score = -0.17, R) >droPer1.super_0 11768957 104 - 11822988 CGGCCGUGGUCAUACACUGCGGUUCGGGCCCAAAUCGUGGCCACUACAUCAGCAUCGUGAAGAGUCACGGCCUCUGGCUGCUGUUCGACGAUGAUAUGGUCGAU---------------- (((((((((((((....((.(((....)))))....))))))))........(((((((((.(((...((((...))))))).))).))))))....)))))..---------------- ( -40.20, z-score = -1.37, R) >dp4.chr2 24565686 104 - 30794189 CGGCCGUGGUCAUACACUGCGGUUCGGGCCCAAAUCGUGGCCACUACAUCAGCAUCGUGAAGAGUCACGGCCUCUGGCUGCUGUUCGACGAUGAUAUGGUCGAU---------------- (((((((((((((....((.(((....)))))....))))))))........(((((((((.(((...((((...))))))).))).))))))....)))))..---------------- ( -40.20, z-score = -1.37, R) >droAna3.scaffold_13340 10982133 104 - 23697760 CCGCCGUGGUCAUUCACUGCGGAUCGGGUCCCAAUCGGGGACACUAUAUCAGCAUUGUCAAGAGCCAUGGGUUGUGGCUGCUCUUCGACGACGAUAUGGUCGAC---------------- ((((.((((....)))).)))).(((.(((((.....)))))((((((((....((((((((((((((.....)))))...)))).))))).))))))))))).---------------- ( -41.40, z-score = -2.04, R) >droEre2.scaffold_4820 998281 104 - 10470090 CCGCUGUGGUCAUCCACUGCGGUUCCGGACCGAAUCGUGGCCAUUAUAUAUCCAUUGUGAAGAGCCACGGACUGUGGCUGCUAUUCGACGAUGAUAUGGUGGAC---------------- ((((.((((....)))).)))).(((((.(((.....)))))....((((((.((((((((.((((((.....))))))....))).)))))))))))..))).---------------- ( -38.50, z-score = -1.90, R) >droYak2.chr3R 19452566 104 + 28832112 CCGCUGUGGUCAUCCACUGCGGUUCCGGACCGAAUCGUGGCCAUUAUAUAUCGAUUGUGAAGAGCCACGGACUGUGGCUGCUAUUCGACGAUGAUAUGGUGGAC---------------- ((((.((((....)))).)))).(((((.(((.....)))))(((((((((((.(((...((((((((.....)))))).))...))))))).)))))))))).---------------- ( -38.80, z-score = -2.04, R) >droSec1.super_0 18962580 104 + 21120651 CCGCUGUGGUUAUCCACUGCGGUUCCGGACCAAAUCGUGGCCAUUAUAUAUCCAUUGUGAAGAGUCACGGACUGUGGCUACUAUUCGACGAUGAUAUGGUGGAC---------------- ((((.((((....)))).)))).(((((.(((.....)))))....((((((.((((((((.((((((.....))))))....))).)))))))))))..))).---------------- ( -37.60, z-score = -2.41, R) >droSim1.chr3R 18414042 104 + 27517382 CCGCUGUGGUUAUCCACUGCGGUUCCGGACCAAAUCGUGGCCAUUAUAUAUCCAUUGUGAAGAGUCACGGACUGUGGCUACUAUUCGACGAUGAUAUGGUGGAC---------------- ((((.((((....)))).)))).(((((.(((.....)))))....((((((.((((((((.((((((.....))))))....))).)))))))))))..))).---------------- ( -37.60, z-score = -2.41, R) >triCas2.ChLG4 10412965 116 - 13894384 UAGCUGUUGUGAUACAUUGCGGAAGCGGACCUAACAGAGGCCACUACAUCAGUAUAGUCAAGAGCCACGGUUUCUGGCUGCUGUUUGACGACGACCAAGUUGACGUAAGUU-UAGUG--- ..((((.((((.......((....))((.(((.....)))))..)))).))))..(((((..(((....)))..)))))((((....(((.((((...)))).))).....-)))).--- ( -33.00, z-score = -0.54, R) >apiMel3.GroupUn 330094239 120 - 399230636 UAGCAGUUGUAAUACAUUGUGGAAGUGGACCUAAUCGAGGACAUUAUAUUUCCAUAGUUAAAAGCCAUGGAUUUUGGUUGCUUUUUGAUGAUGACAUGGUUGAUGUAAGUCAUUAUAAUU .(((...........((((((((((((..(((.....)))......))))))))))))....(((((.......))))))))...(((((((.((((.....))))..)))))))..... ( -26.00, z-score = -0.58, R) >consensus CCGCUGUGGUCAUACACUGCGGUUCGGGACCGAAUCGUGGCCAUUAUAUAAGCAUUGUGAAGAGUCACGGACUGUGGCUGCUGUUCGACGAUGAUAUGGUCGAC________________ (((((((.(((...............((.(((.....)))))............(((...(((((((.......))))).))...)))....)))))))))).................. (-16.53 = -15.55 + -0.98)
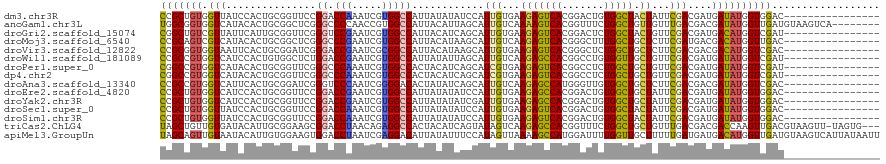

| Location | 18,601,842 – 18,601,932 |
|---|---|
| Length | 90 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Shannon entropy | 0.32988 |
| G+C content | 0.47137 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -15.14 |
| Energy contribution | -13.98 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18601842 90 + 27905053 ---------------------------AAAAUCGAGGCUUCCACAAUCGAGGACUUCUAUGGUCUGACCUCGGACAUACACAAGUCUUCGGAGACCGGCUACAUACUGUUCUAUCAG ---------------------------......(.((((.......((((((((((.....(((((....)))))......)))))))))).....)))).)...(((......))) ( -22.70, z-score = -1.27, R) >anoGam1.chr3L 3957868 117 - 41284009 AUCAAUUUUCCCUCUUUCCAUCUGCAGAAAAUUGAAGCUUCCACCAUCGAGGACUUUUACGGGUUGACAUCGGACAUACAAAAGUCUUCGGAAACGGGUUACAUUCUGUUCUACCAG .(((((((((................)))))))))...........((((((((((((....(((.......)))....)))))))))))).((((((......))))))....... ( -24.89, z-score = -0.59, R) >droGri2.scaffold_15074 3396721 90 + 7742996 ---------------------------AAGAUUGAGGCCUCAACCAUUGAGGAUUUUUAUGGCCUCACCUCCGAUAUACACAAAUCCUCGGAGACCGGUUACAUUCUAUUCUAUCAA ---------------------------.(((.((..(((........(((((..........))))).((((((.............))))))...)))..)).))).......... ( -20.72, z-score = -1.47, R) >droMoj3.scaffold_6540 1107924 90 - 34148556 ---------------------------AAAAUCGAGGCUUCGACCAUUGAGGACUUCUACGGCCUCACCUCGGACAUACACAAGUCCUCCGAAACGGGCUAUAUAUUGUUCUAUCAG ---------------------------......((((..........(((((.((.....)))))))))))(((((......(((((........)))))......)))))...... ( -20.90, z-score = -1.23, R) >droVir3.scaffold_12822 3683287 90 - 4096053 ---------------------------AAGAUUGAGGCCUCGACCAUCGAGGACUUUUAUGGCCUCACCUCGGAUAUACACAAGUCCUCGGAAACCGGCUACAUUCUGUUCUAUCAA ---------------------------.(((.((.((((.........((((((((.((((.((.......)).))))...))))))))(....).)))).)).))).......... ( -25.80, z-score = -2.18, R) >droWil1.scaffold_181089 10937165 90 - 12369635 ---------------------------AAAAUUGAAGCCUCUACCAUUGAGGACUUCUAUGGCCUCACCUCGGACAUACACAAGUCCUCGGAAACGGGUUAUAUAUUGUUCUAUCAG ---------------------------......((((((((.......)))).))))(((((((.......((((........)))).((....))))))))).............. ( -25.50, z-score = -2.95, R) >droPer1.super_0 11769061 90 - 11822988 ---------------------------AAGAUUGAGGCCUCCACCAUCGAGGACUUUUACGGCCUCACCUCGGACAUACACAAGUCCUCGGAGACGGGCUAUAUAUUAUUCUAUCAG ---------------------------.(((.((((((((((........))).......)))))))((..((((........)))).((....))))...........)))..... ( -23.71, z-score = -1.73, R) >dp4.chr2 24565790 90 - 30794189 ---------------------------AAGAUUGAGGCCUCCACCAUCGAGGACUUUUACGGCCUCACCUCGGACAUACACAAGUCCUCGGAGACGGGCUAUAUAUUAUUCUAUCAG ---------------------------.(((.((((((((((........))).......)))))))((..((((........)))).((....))))...........)))..... ( -23.71, z-score = -1.73, R) >droAna3.scaffold_13340 10982237 90 - 23697760 ---------------------------AAGAUUGAAGCCUCCACCAUCGAGGACUUCUACGGCCUGACCUCGGACAUACACAAGUCCUCGGAGACGGGCUAUAUUCUAUUCUAUCAG ---------------------------.(((.((.(((((....(..(((((((((.....(.(((....))).)......)))))))))..)..))))).)).))).......... ( -25.50, z-score = -1.98, R) >droEre2.scaffold_4820 998385 90 - 10470090 ---------------------------AAAAUCGAGGCUUCCACCAUCGAGGACUUCUACGGUUUGACCUCGGACAUACACAAGUCCUCGGAGACGGGCUACAUACUGUUCUAUCAG ---------------------------......(.(((..((..(..(((((((((.....(((((....)))))......)))))))))..)..))))).)...(((......))) ( -24.10, z-score = -1.40, R) >droYak2.chr3R 19452670 90 + 28832112 ---------------------------AAAAUCGAGGCUUCCACCAUCGAGGACUUCUACGGCCUGACCUCCGACAUACACAAGUCCUCGGAAACGGGCUACAUACUGUUCUAUCAG ---------------------------....((((((......)).))))((((......((((((...(((((...((....))..)))))..)))))).......))))...... ( -22.42, z-score = -1.66, R) >droSec1.super_0 18962684 90 + 21120651 ---------------------------AAAAUCGAGGCUUCCACCAUCGAGGAUUUCUACGGUCUGACCUCGGACAUACACAAGUCCUCGGAGACGGGCUACAUACUGUUCUAUCAG ---------------------------......(.(((..((..(..(((((((((.....(((((....)))))......)))))))))..)..))))).)...(((......))) ( -24.50, z-score = -1.39, R) >droSim1.chr3R 18414146 90 + 27517382 ---------------------------AAAAUCGAGGCUUCCACCAUCGAGGACUUCUACGGUCUGACCUCGGACAUACACAAGUCCUCGGAGACGGGCUACAUACUGUUCUAUCAG ---------------------------......(.(((..((..(..(((((((((.....(((((....)))))......)))))))))..)..))))).)...(((......))) ( -26.80, z-score = -2.08, R) >triCas2.ChLG4 10413121 94 - 13894384 -----------------------AUAGAAAAUAGACGCUUCAACAAUAGAAGAUUUCUACGGCCUCACGUCUGAUACUCAAAAGUCUUCAGAAACAGGCUACAUCCUUUUUUACCAG -----------------------.(((((((((((..((((.......))))...)))).(((((....(((((.(((....)))..)))))...)))))......))))))).... ( -20.50, z-score = -3.04, R) >consensus ___________________________AAAAUUGAGGCUUCCACCAUCGAGGACUUCUACGGCCUCACCUCGGACAUACACAAGUCCUCGGAGACGGGCUACAUACUGUUCUAUCAG .................................((((((((.......)))).))))...((((.......((((........))))..(....).))))................. (-15.14 = -13.98 + -1.16)

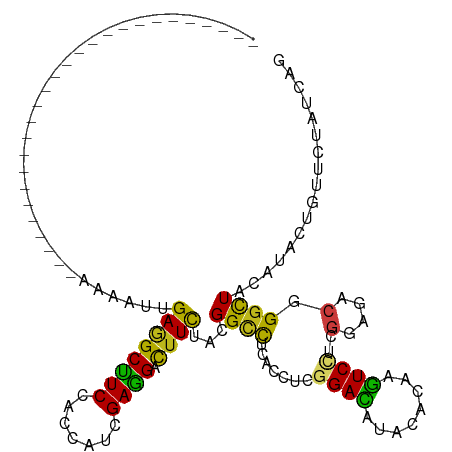

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:36:00 2011