| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,311,622 – 8,311,738 |
| Length | 116 |
| Max. P | 0.721451 |

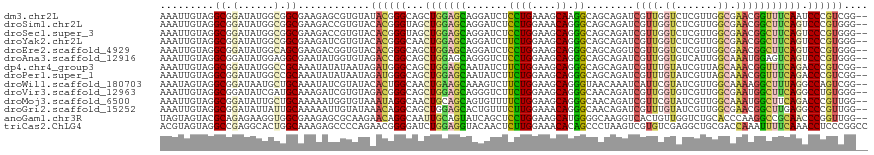
| Location | 8,311,622 – 8,311,738 |
|---|---|
| Length | 116 |
| Sequences | 14 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Shannon entropy | 0.57199 |
| G+C content | 0.53440 |
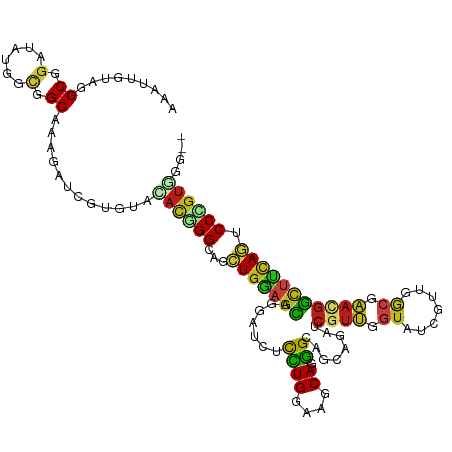
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -18.68 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8311622 116 - 23011544 AAAUUGUAGGCGGAUAUGGCGGCGAAGAGCGUGUAUACGGGCAGCUGGAGCAGGAUCUCCUGAAAGCAAGGCAGCAGAUCGUUGGUCUCGUUGGCGAACGGUUUCAAUCCCGUCGG-- ........(((((.....((.((.....((.((....)).)).(((....((((....))))..)))...)).))((((((((.(((.....))).)))))))).....)))))..-- ( -39.10, z-score = -0.58, R) >droSim1.chr2L 8099546 116 - 22036055 AAAUUGUAGGCGGAUAUGGCGGCGAAGACCGUGUACACGGGUAGCUGGAGCAGGAUCUCCUGGAAACAGGGCAGCAGAUCGUUGGUCUCGUUGGCGAACGGCUUCAGUCCCGUGGG-- .............(((((((......).)))))).((((((..((((((((..(((((((((....)))).....)))))(((.(((.....))).))).))))))))))))))..-- ( -48.40, z-score = -2.72, R) >droSec1.super_3 3807786 116 - 7220098 AAAUUGUAGGCGGAUAUGGCGGCGAAGACCGUGUACACGGGUAGCUGGAGCAGGAUCUCCUGGAAGCAGGGCAGCAGAUCGUUGGUCUCGUUGGCGAACGGCUUCAGUCCCGUGGG-- .............(((((((......).)))))).((((((..((((((((..(((((((((....)))).....)))))(((.(((.....))).))).))))))))))))))..-- ( -45.90, z-score = -1.73, R) >droYak2.chr2L 10949425 116 - 22324452 AAAUUGUAGGCGGAUAUGGCGGCGAAGAUCGUGUACACGGGCAACUGGAGCAGGAUCUUCUGGAAGCAGGGCAGCAGAUCGUUGGUCUCGUUGGCGAACGGCUUCAGUCCCGUGGG-- ..............(((((.((((((((((.(((.(.(((....)))).))).)))))))..(((((..(.((((((((.....)))).)))).).....))))).))))))))..-- ( -43.60, z-score = -1.85, R) >droEre2.scaffold_4929 17221168 116 - 26641161 AAAUUGUAGGCGGAUAUGGCAGCGAAGACGGUGUACACGGGCAGCUGGAGCAGGAUCUCCUGGAAGCAGGGCAGCAGGUCGUUGGUCUCGUUGGCGAACGGCUUCAGUCCCGUGGG-- ..................(((.((....)).))).((((((..((((((((..(((((((((....)))).....)))))(((.(((.....))).))).))))))))))))))..-- ( -43.90, z-score = -1.22, R) >droAna3.scaffold_12916 10226720 116 + 16180835 AAAUUGUAGGCGGAUAUGGAGGCGAAUAUGGUGUAGACCGGCAGCUGGAGCAGGGUCUCCUGGAAGCAGGGCAGCAGAUCGUUGGUGUCAUUGGCAAAUGGAGUCAGUCCCGUGGG-- .........((((((((...(((((...((.((((((((.((.......))..)))))((((....))))))).))..))))).)))))((((((.......))))))..)))...-- ( -34.10, z-score = 0.34, R) >dp4.chr4_group3 7839671 116 + 11692001 AAAUUGUAGGCGGAUAUGGCCGCAAAUAUAUAAUAGAUGGGCAGCUGGAGCAAUAUCUUCUGGAAGCAGGGCAGCAGAUCGUUUGUAUCGUUAGCAAACGGUUUCAGACCCGUCGG-- ..((((((.((((......)))).....)))))).(((((((.((((((......))(((((....)))))))))(((((((((((.......)))))))))))..).))))))..-- ( -38.90, z-score = -2.83, R) >droPer1.super_1 4950205 116 + 10282868 AAAUUGUAGGCGGAUAUGGCCGCAAAUAUAUAAUAGAUGGGCAGCUGGAGCAAUAUCUUCUGGAAGCAGGGCAGCAGAUCGUUUGUAUCGUUAGCAAACGGUUUCAGACCCGUCGG-- ..((((((.((((......)))).....)))))).(((((((.((((((......))(((((....)))))))))(((((((((((.......)))))))))))..).))))))..-- ( -38.90, z-score = -2.83, R) >droWil1.scaffold_180703 423656 116 - 3946847 AAAUAGUAGGCGGAUAAUGCUGCAAAUAUCGUAUACACUGGCAACUGAAGCAAAGUCUUCUGGAAGCAGGGUAACAAAUCAUUCGUAUCGUUGGCAAAAGGCUUUAGGCCAGUCGG-- ..(..(((.((((......))))...)))..)...(((((((..(((((((........(((....)))(.((((..............)))).).....))))))))))))).).-- ( -27.54, z-score = -0.62, R) >droVir3.scaffold_12963 16066736 116 - 20206255 AAAUUGUAGGCGGAUAUCGAUGCAAAGAUCGUGUAGACGGGCAGCUGGAGCAGGGUCUUCUGGAAGCAGGGCAACAGAUCGUUGGUGUCGUUGGCGAAUGGCUUCAGGCCUGUGGG-- ...(..(((((.((((((((((..((((((.(((...(((....)))..))).))))))(((....)))(....)....))))))))))(..(((.....))).)..)))))..).-- ( -37.00, z-score = -0.71, R) >droMoj3.scaffold_6500 5525378 116 - 32352404 AAAUUGUAGGCGGAUAUUGCUGCAAAAAUGGUGUAAAUAGGCAACUGCAGCAGUGUUUUCUGGAAGCAGGGCAACAGAUCGUUCGUAUCGUUGGCAAAUGGCUUCAGACCCGUUGG-- ........((((((((((((((((.......(((......)))..)))))))))))..((((.((((..(.((((.((((....).))))))).).....)))))))).)))))..-- ( -34.90, z-score = -1.25, R) >droGri2.scaffold_15252 13503864 116 - 17193109 AAAUUGUAGGCGGAUAUUAUUGCAAAAAUUGUAUAAACAGGCAGCUGGAGCACUGUUUCUUGGAAACAGGGCAACAGAUCGUUUGUAUCGUUGGCGAACGGCUUGAGGCCCGUUGG-- ...((((((..........))))))..........(((.(((..((.((((.((((((((((....))))).)))))..(((((((.......))))))))))).))))).)))..-- ( -34.90, z-score = -1.74, R) >anoGam1.chr3R 2787790 116 - 53272125 UAGUAGUACGCAGAGAAGGUGGCGAAGAGCGCAAGAACAGGCAAUUGCAGUAUCAGCUCCUGGAAGCAUGGGGCAAGGUCACUGUUGGUCUGCACCCAAGGCCGCAACCCGGUUGG-- .................(((((((.....))).....(((((....((((((((.(((((((....)).)))))..))).)))))..)))))))))...(((((.....)))))..-- ( -35.10, z-score = 0.76, R) >triCas2.ChLG4 4956691 118 - 13894384 ACGUAGUAGGCCGAGGCACUGGCAAAGAGCCCCAGAACGGGGAUCUGGAGGUACAACUCUUGGAAACACAGCCCUAAGUCGUGUCGAGGCUGCGACCAAAUUUUCAAACCUCCCGGCC ........(((((..((....))..(((.((((.....)))).)))((((((........((....)).........(((((((....)).)))))...........))))))))))) ( -38.40, z-score = -0.36, R) >consensus AAAUUGUAGGCGGAUAUGGCGGCAAAGAUCGUGUACACGGGCAGCUGGAGCAGGAUCUCCUGGAAGCAGGGCAGCAGAUCGUUGGUAUCGUUGGCGAACGGCUUCAGUCCCGUGGG__ .........((.(......).))............((((((...(((((((.......((((....)).))........((((.((.......)).))))))))))).)))))).... (-18.68 = -18.57 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:43 2011