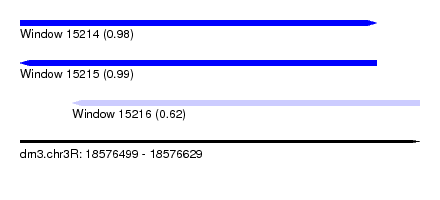
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,576,499 – 18,576,629 |
| Length | 130 |
| Max. P | 0.992153 |

| Location | 18,576,499 – 18,576,615 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.32 |
| Shannon entropy | 0.46149 |
| G+C content | 0.47416 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.89 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
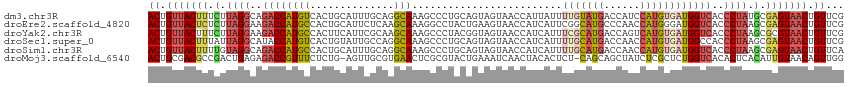
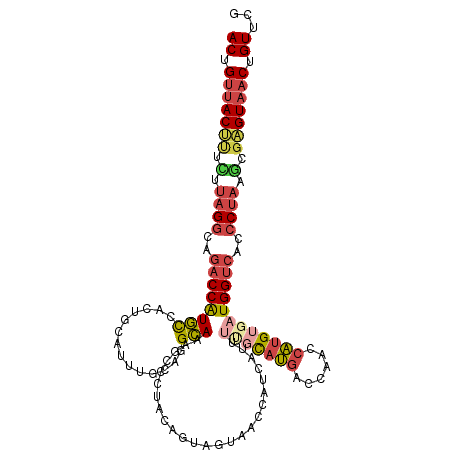
>dm3.chr3R 18576499 116 + 27905053 ACUGUUACUUUCUUAGGCAGACCAUGUCACUGCAUUUGCAGGCAAAGCCCUGCAGUAGUAACCAUUAUUUUGUAUGACCAUCCAUGUGAUGGUCACCCUAUGCGAGUAACUGUUCG ((.(((((((.(.((((..(((((.((.(((((...((((((......))))))))))).)).......(..((((......))))..))))))..)))).).))))))).))... ( -36.80, z-score = -3.15, R) >droEre2.scaffold_4820 972150 116 - 10470090 ACUGUUACUCUCUUAGGAAGACCAUGCCACUGCAUUCUCAAGCAAAGGCCUACUGAAGUAACCAUCAUUCGGCAUGCCCAACCAUGGGAUGGUCACCCUAAGCGAGUAACUGUUCG ((.(((((((.((((((..((((((.(((.(((........)))..(((...(((((..........)))))...)))......))).))))))..)))))).))))))).))... ( -43.20, z-score = -5.00, R) >droYak2.chr3R 19425756 116 + 28832112 ACUGUUACUUUCUUAGGAAGACCAUGCCACUUCAUUCGCAAGCAAAGCCCUACGGUAGUAACCAUCAUUUCGCAUGACCAGUCAUGUGAUGGUCACCCUAAGCGCGUAACUGUUCG ((.(((((...((((((..((((((((..........))).............(((....)))......((((((((....)))))))))))))..))))))...))))).))... ( -33.10, z-score = -2.97, R) >droSec1.super_0 18932977 116 + 21120651 ACUGUUACUUUAUUAGGCAUACCAUGUCACUGUAUUGCCAGGCAAAGCCCUGCAGUAGUAACCAUCAUUUUGCAUGACCAACCAUGUGAUGGCCACCCUAAGCGAGUAACUGUUCG ((.(((((((..(((((...............((((((..(((...)))..))))))((..(((((.....(((((......))))))))))..)))))))..))))))).))... ( -31.30, z-score = -1.52, R) >droSim1.chr3R 18387408 116 + 27517382 ACUGUUACUUUUGUAGGCAGACCAUGCCACUGCAUUUGCAGGCAAAGCCCUGCAGUAGUAACCAUCAUUUUGCAUGACCAACCAUGUGAUGGUCACCCUAAGCGAGUAACUGUUCA ((.(((((((...((((..((((((...(((((...((((((......)))))))))))............(((((......))))).))))))..))))...))))))).))... ( -38.40, z-score = -2.82, R) >droMoj3.scaffold_6540 1086513 114 - 34148556 ACUGCGACGCCGACUGAGAGACCGUUUCUCUG-AGUUGCGUGAACUCGCGUACUGAAAUCAACUACACUCU-CAGCAGCUAUCUCGCUCUGGUCACACUCACAUUGUAACAGUUGG ...((((((((((((.(((((.....))))).-))))).)))...))))..........(((((.......-(((.(((......))))))((.(((.......))).))))))). ( -31.70, z-score = -0.67, R) >consensus ACUGUUACUUUCUUAGGCAGACCAUGCCACUGCAUUUGCAGGCAAAGCCCUACAGUAGUAACCAUCAUUUUGCAUGACCAACCAUGUGAUGGUCACCCUAAGCGAGUAACUGUUCG ((.(((((((.(.((((..((((((((..............))).........................(((((((......))))))))))))..)))).).))))))).))... (-16.69 = -17.89 + 1.20)
| Location | 18,576,499 – 18,576,615 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.32 |
| Shannon entropy | 0.46149 |
| G+C content | 0.47416 |
| Mean single sequence MFE | -41.53 |
| Consensus MFE | -21.76 |
| Energy contribution | -23.27 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18576499 116 - 27905053 CGAACAGUUACUCGCAUAGGGUGACCAUCACAUGGAUGGUCAUACAAAAUAAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGACAUGGUCUGCCUAAGAAAGUAACAGU ......((((((..(.(((((((((((((.....)))))))))...........((((((.(((((((......))))))...).))))))........)))).)..))))))... ( -40.90, z-score = -3.39, R) >droEre2.scaffold_4820 972150 116 + 10470090 CGAACAGUUACUCGCUUAGGGUGACCAUCCCAUGGUUGGGCAUGCCGAAUGAUGGUUACUUCAGUAGGCCUUUGCUUGAGAAUGCAGUGGCAUGGUCUUCCUAAGAGAGUAACAGU ......(((((((.(((((((.((((((.((((.(((.((....)).))).))))((((....))))(((.((((........)))).))))))))).))))))).)))))))... ( -50.90, z-score = -4.95, R) >droYak2.chr3R 19425756 116 - 28832112 CGAACAGUUACGCGCUUAGGGUGACCAUCACAUGACUGGUCAUGCGAAAUGAUGGUUACUACCGUAGGGCUUUGCUUGCGAAUGAAGUGGCAUGGUCUUCCUAAGAAAGUAACAGU ......(((((...(((((((((((((((((((((....))))).....))))))))))......((((((.((((.((.......)))))).))))))))))))...)))))... ( -42.40, z-score = -3.55, R) >droSec1.super_0 18932977 116 - 21120651 CGAACAGUUACUCGCUUAGGGUGGCCAUCACAUGGUUGGUCAUGCAAAAUGAUGGUUACUACUGCAGGGCUUUGCCUGGCAAUACAGUGACAUGGUAUGCCUAAUAAAGUAACAGU ......((((((.((....((((((((((((((((....))))).....)))))))))))...)).((((..((((((.((......)).)).)))).)))).....))))))... ( -37.20, z-score = -1.44, R) >droSim1.chr3R 18387408 116 - 27517382 UGAACAGUUACUCGCUUAGGGUGACCAUCACAUGGUUGGUCAUGCAAAAUGAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGGCAUGGUCUGCCUACAAAAGUAACAGU ......((((((....(((((((((((((((((((....))))).....)))))))))))...(((((.((.(((((((....)))..)))).))))))))))....))))))... ( -41.00, z-score = -2.14, R) >droMoj3.scaffold_6540 1086513 114 + 34148556 CCAACUGUUACAAUGUGAGUGUGACCAGAGCGAGAUAGCUGCUG-AGAGUGUAGUUGAUUUCAGUACGCGAGUUCACGCAACU-CAGAGAAACGGUCUCUCAGUCGGCGUCGCAGU ....((((((((.......))))).))).((((....(((((((-((((....(((..(((.(((..(((......))).)))-.)))..)))...))))))).)))).))))... ( -36.80, z-score = -0.77, R) >consensus CGAACAGUUACUCGCUUAGGGUGACCAUCACAUGGUUGGUCAUGCAAAAUGAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGACAUGGUCUGCCUAAGAAAGUAACAGU ......((((((....((((..((((((..(((((....)))))..........((((((...(((((......)))))......))))))))))))..))))....))))))... (-21.76 = -23.27 + 1.50)
| Location | 18,576,516 – 18,576,629 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.63 |
| Shannon entropy | 0.46554 |
| G+C content | 0.46322 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -18.25 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18576516 113 - 27905053 -AAAGUAAAACUCG---GCGAACAGUUACUCGCAUAGGGUGACCAUCACAUGGAUGGUCAUACAAAAUAAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGACAUGGUCUG -...(((..((((.---((((........))))...))))(((((((.....))))))).))).........((((((.(((((((......))))))...).))))))........ ( -36.00, z-score = -2.63, R) >droEre2.scaffold_4820 972167 114 + 10470090 AAAAGUAAAACUCU---GCGAACAGUUACUCGCUUAGGGUGACCAUCCCAUGGUUGGGCAUGCCGAAUGAUGGUUACUUCAGUAGGCCUUUGCUUGAGAAUGCAGUGGCAUGGUCUU .........(((((---((((........))))..)))))((((((.((((.(((.((....)).))).))))((((....))))(((.((((........)))).))))))))).. ( -37.10, z-score = -1.47, R) >droYak2.chr3R 19425773 114 - 28832112 AAAAGUAAAACUCU---GCGAACAGUUACGCGCUUAGGGUGACCAUCACAUGACUGGUCAUGCGAAAUGAUGGUUACUACCGUAGGGCUUUGCUUGCGAAUGAAGUGGCAUGGUCUU ..(((((((.((((---(((...(((.....)))...((((((((((((((((....))))).....)))))))))))..))))))).)))))))((.(......).))........ ( -35.60, z-score = -1.87, R) >droSec1.super_0 18932994 113 - 21120651 -AAAGUAAAACUCA---GCGAACAGUUACUCGCUUAGGGUGGCCAUCACAUGGUUGGUCAUGCAAAAUGAUGGUUACUACUGCAGGGCUUUGCCUGGCAAUACAGUGACAUGGUAUG -...(((...((.(---((((........))))).))((((((((((((((((....))))).....)))))))))))..(((.((((...)))).))).))).............. ( -33.30, z-score = -0.94, R) >droSim1.chr3R 18387425 113 - 27517382 -AAAGUAAAACUCG---GUGAACAGUUACUCGCUUAGGGUGACCAUCACAUGGUUGGUCAUGCAAAAUGAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGGCAUGGUCUG -...(((...((.(---((((........))))).))((((((((((((((((....))))).....)))))))))))..))).(((((.(((((((....)))..)))).))))). ( -38.40, z-score = -2.24, R) >droMoj3.scaffold_6540 1086530 113 + 34148556 --AAAUAAACUCCAACUGCCAACUGUUACAAUGUGAGUGUGACCAGAGCGAGA-UAGCUGCUGAGAGUGUAGUUGAUUUCAGUACGCGAGUUCACGCAACU-CAGAGAAACGGUCUC --...................((((((....(.((((((((..((((((....-..))).)))...(((((.(((....)))))))).......))).)))-)).)..))))))... ( -26.40, z-score = 0.49, R) >consensus _AAAGUAAAACUCA___GCGAACAGUUACUCGCUUAGGGUGACCAUCACAUGGUUGGUCAUGCAAAAUGAUGGUUACUACUGCAGGGCUUUGCCUGCAAAUGCAGUGACAUGGUCUG ........................(((((........(((((((((((((((......)))).....)))))))))))...(((((......))))).......)))))........ (-18.25 = -19.17 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:55 2011