
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,555,810 – 18,555,879 |
| Length | 69 |
| Max. P | 0.899513 |

| Location | 18,555,810 – 18,555,879 |
|---|---|
| Length | 69 |
| Sequences | 12 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 87.42 |
| Shannon entropy | 0.27984 |
| G+C content | 0.52369 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.91 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18555810 69 - 27905053 GCUAUAUACUUA---UUCUACAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- ............---.....((((.((.(((((((((((((....)))).))))).))))))..))))....- ( -22.50, z-score = -1.50, R) >droSim1.chr3R 18366772 69 - 27517382 GCUAUAUACUUA---UUCUACAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- ............---.....((((.((.(((((((((((((....)))).))))).))))))..))))....- ( -22.50, z-score = -1.50, R) >droSec1.super_0 18912396 69 - 21120651 GCUAUAUACUUA---UUCUACAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- ............---.....((((.((.(((((((((((((....)))).))))).))))))..))))....- ( -22.50, z-score = -1.50, R) >droYak2.chr3R 19407319 69 - 28832112 GCUAUAUACUUA---UUCUACAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- ............---.....((((.((.(((((((((((((....)))).))))).))))))..))))....- ( -22.50, z-score = -1.50, R) >droEre2.scaffold_4820 952723 69 + 10470090 GCUAUAUACUUA---UUCUACAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- ............---.....((((.((.(((((((((((((....)))).))))).))))))..))))....- ( -22.50, z-score = -1.50, R) >droAna3.scaffold_13340 10938372 70 + 23697760 GCUAUAUACUUAG--GUCUAGAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- .(((......)))--..(((((...((.(((((((((((((....)))).))))).))))))..)))))...- ( -24.00, z-score = -1.12, R) >dp4.chr2 24515767 71 + 30794189 GCUAUAUACUUAGA-AUCUAAAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- .(((......))).-..(((((...((.(((((((((((((....)))).))))).))))))..)))))...- ( -23.70, z-score = -1.65, R) >droWil1.scaffold_181089 10893382 72 + 12369635 UCUAUAUACUUAUAUAUCUAAAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- .................(((((...((.(((((((((((((....)))).))))).))))))..)))))...- ( -23.30, z-score = -2.10, R) >droVir3.scaffold_13047 6008737 70 + 19223366 GCUAGAUGCU--UAUGUCUAAAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- (((((((((.--...(((........)))((((((((((((....)))).))))).)))..)))))))))..- ( -29.90, z-score = -2.94, R) >droMoj3.scaffold_6540 23696587 72 + 34148556 UCUAGAUGCUUAUAUGUCUAAAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- .((((((((......(((........)))((((((((((((....)))).))))).)))..))))))))...- ( -28.50, z-score = -2.81, R) >droGri2.scaffold_14906 4478506 70 - 14172833 GCUAGAUGCU--UAUGUCUAAAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU- (((((((((.--...(((........)))((((((((((((....)))).))))).)))..)))))))))..- ( -29.90, z-score = -2.94, R) >anoGam1.chrX 7926858 65 - 22145176 ----UAGACUAG---GUCUAAAGCACAACGUCGGGCAGAGCGUGUGCCCUGGCCUAGUCUCUUA-ACGAUGCG ----.(((((((---(((....(((((.((((.....)).)))))))...))))))))))....-........ ( -25.30, z-score = -2.57, R) >consensus GCUAUAUACUUA___GUCUAAAGAACGACGUCAGGCAGGUCGUGCGGCCUUGCCUAGGCGCGCAUUUGGUUU_ .........................((.(((((((((((((....))))).)))).))))))........... (-18.72 = -18.91 + 0.19)
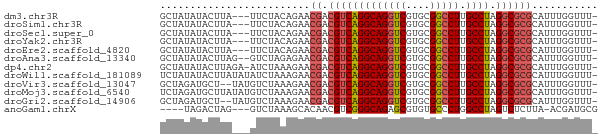
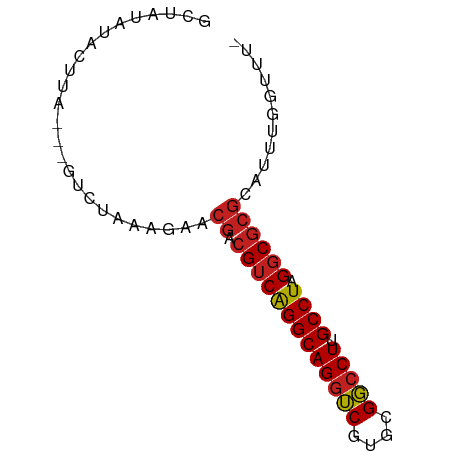

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:53 2011