| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,490,721 – 18,490,820 |
| Length | 99 |
| Max. P | 0.964520 |

| Location | 18,490,721 – 18,490,820 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.96 |
| Shannon entropy | 0.53065 |
| G+C content | 0.60866 |
| Mean single sequence MFE | -32.31 |
| Consensus MFE | -18.16 |
| Energy contribution | -17.52 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
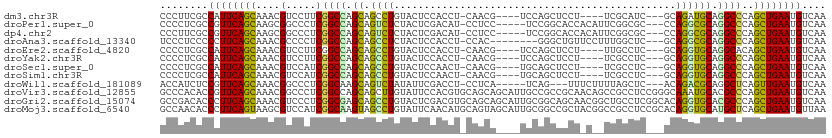
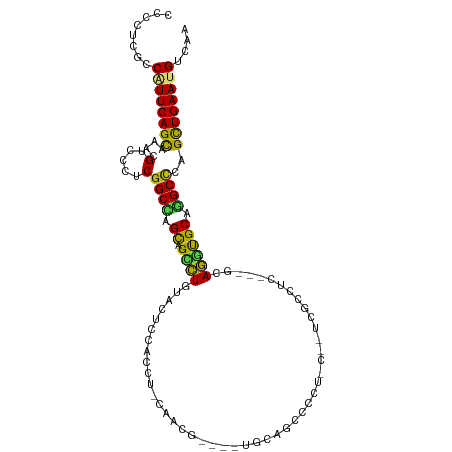
>dm3.chr3R 18490721 99 + 27905053 CCCUUCGCCAUUCAGCAAACGUCCUUCGGCCAGCAGCCUGUACUCCACCU-CAACG----UCCAGCUCCU----UCGCAUC---GCAGAUGCAGGCCCAGCUGAAUGUCAA ......(.((((((((....(.....)((((((.(((.((.((.......-....)----).))))).))----..(((((---...))))).))))..)))))))).).. ( -27.30, z-score = -2.54, R) >droPer1.super_0 3918732 102 - 11822988 CCCCUCGCCGUUCAGCAAGCGGCCCUCGGCCAGCAGUCUCUACUCGACAU-CCUCC-----UCCGGCACCACAUUCGGCGC---CCAGGCGCAGGCCCAGCUGAAUGUCAA ......(.((((((((..((((((...)))).)).(((.......)))..-(((.(-----.(((((.((......)).))---)..)).).)))....)))))))).).. ( -34.20, z-score = -1.52, R) >dp4.chr2 12563336 102 + 30794189 CCCUUCGCCGUUCAGCAAGCGGCCCUCGGCCAGCAGUCUCUACUCGACAU-CCUCC-----UCCGGCACCACAUUCGGCGC---CCAGGCGCAGGCCCAGCUGAAUGUCAA ......(.((((((((..((((((...)))).)).(((.......)))..-(((.(-----.(((((.((......)).))---)..)).).)))....)))))))).).. ( -34.20, z-score = -1.55, R) >droAna3.scaffold_13340 15564511 99 - 23697760 UCCCUCCCCCUUCAGCAAACGCCCCUCGGCCAGCAGCCUCUACUCCACCU-CCAC--------GGGCUGUUCCUUUGGCUC---GCAGGCGCAGGCCCAGCUGAAUGUCAA ..........((((((....(((((((((((((((((((...........-....--------))))))).....)))).)---).)))....)))...))))))...... ( -29.86, z-score = -0.78, R) >droEre2.scaffold_4820 884648 99 - 10470090 CCCCUCGCCAUUCAGCAAACGUCCUUCGGCCAGCAGCCUGUACUCCACCU-CAACG----UCCAGCUCCU----UUGCCUC---GCAGGUGCAGGCACAGCUGAAUGUCAA ......(.((((((((...((.....))(((.(((.(((((.....((..-....)----)...((....----..))...---)))))))).)))...)))))))).).. ( -28.80, z-score = -2.02, R) >droYak2.chr3R 19341221 99 + 28832112 CCCCUCGCCAUUCAGCAAACGUCCUUCGGCCAGCAGCCUGUACUCCACCU-CAACG----UCCAGCUCCU----UCGCCUC---GCAGGUGCAGGCCCAGCUGAAUGUCAA ......(.((((((((....(.....)((((.(((.(((((.....((..-....)----)...((....----..))...---)))))))).))))..)))))))).).. ( -28.70, z-score = -2.47, R) >droSec1.super_0 18847941 99 + 21120651 CCCCUCGCCAUUCAGCAAACGUCCAUCGGCCAGCAGCCUGUACUCCAACU-CAACG----UGCAGCUCCU----UCGCCUC---GCAGGUGCAGGCCCAGCUGAAUGUCAA ......(.((((((((....(.....)((((((.(((.(((((.......-....)----))))))).))----.((((..---...))))..))))..)))))))).).. ( -30.40, z-score = -2.26, R) >droSim1.chr3R 18300591 99 + 27517382 CCCCUCGCCAUUCAGCAAACGUCCAUCGGCCAGCAGCCUGUACUCCAACU-CAACG----UGCAGCUCCU----UCGCCUC---GCAGGUGCAGGCCCAGCUGAAUGUCAA ......(.((((((((....(.....)((((((.(((.(((((.......-....)----))))))).))----.((((..---...))))..))))..)))))))).).. ( -30.40, z-score = -2.26, R) >droWil1.scaffold_181089 6166214 99 - 12369635 ACCAUCUCCGUUCAGCAAACGGCCCUCGGCAAGCAGUCUAUAUUCGACCU-CCUCA-----UCAG---UUUCUUUUAGCUC---ACAGACGCAGGCUCAGUUGAAUGUCAA ........((((((((.....(((...))).(((.((((......((...-..)).-----..((---((......)))).---..))))....)))..)))))))).... ( -18.60, z-score = -0.32, R) >droVir3.scaffold_12855 7773397 111 - 10161210 GCCCACACCGUUCAGCAAACGGCCCUCGGCCAGCAGCUUGUAUUCCACGUGCAGCAGCAUUGCCGCCGCAACAGCCGCCUCCGGGCAAAUGCACGCCCAGCUGAAUGUCAA ........((((((((...((((...((((..(((((((((((.....)))))..)))..))).)))).....)))).....((((........)))).)))))))).... ( -38.80, z-score = -1.28, R) >droGri2.scaffold_15074 5348139 111 - 7742996 GCCGACACCCUUCAGCAAACGUCCCUCGGCGAGCAGCCUGUACUCGACGUGCAGCAGCAUUGCGGCAGCAACGGCUGCCUCGGCACAGGUGCACGCCCAGCUGAAUGUCAA ...((((...((((((....(.....)((((.(((.(((((.(.(((.((((....))))...((((((....)))))))))).)))))))).))))..)))))))))).. ( -47.00, z-score = -1.90, R) >droMoj3.scaffold_6540 34072142 111 + 34148556 GCCAACACCCUUCAGUAAGCGUCCAUCGGCAAGUAGCCUGUAUUCAACAUGCAGUAGCAUUGCGGCCGCUACGGCCGCCUCCGCACAGGUGCAUGCUCAGCUGAAUGUUAA ...((((...((((((.(((((.((((.((.....((((((((.....))))))..))...(((((((...)))))))....))...)))).)))))..)))))))))).. ( -39.50, z-score = -2.04, R) >consensus CCCCUCGCCAUUCAGCAAACGUCCCUCGGCCAGCAGCCUGUACUCCACCU_CAACG____UGCAGCCCCU_C__UCGCCUC___GCAGGUGCAGGCCCAGCUGAAUGUCAA ........((((((((....(.....)((((.((.((((...............................................)))))).))))..)))))))).... (-18.16 = -17.52 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:49 2011