| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,465,012 – 18,465,126 |
| Length | 114 |
| Max. P | 0.972091 |

| Location | 18,465,012 – 18,465,126 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 56.63 |
| Shannon entropy | 0.86076 |
| G+C content | 0.35276 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -5.24 |
| Energy contribution | -5.37 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 2.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.972091 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
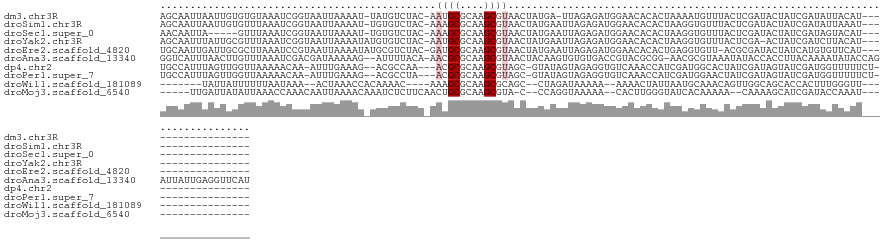
>dm3.chr3R 18465012 114 + 27905053 AGCAAUUAAUUGUGUGUAAAUCGGUAAUUAAAAU-UAUGUCUAC-AAUGCGCAAGCGUAACUAUGA-UUAGAGAUGGAACACACUAAAAUGUUUACUCGAUACUAUCGAUAUUACAU------------------ .((((....))))(((((((((((((.....(((-((((..(((-...((....)))))..)))))-)).(((...(((((........))))).))).....))))))).))))))------------------ ( -24.30, z-score = -1.76, R) >droSim1.chr3R 18277230 115 + 27517382 AGCAAUUAAUUGUGUUUAAAUCGGUAAUUAAAAU-UGUGUCUAC-AAAGCGCAAGCGUAACUAUGAAUUAGAGAUGGAACACACUAAGGUGUUUACUCGAUACUAUCGAUAUUAAAU------------------ .((((....))))(((((((((((((........-.(((((...-...((....))..............(((...((((((......)))))).))))))))))))))).))))))------------------ ( -25.30, z-score = -1.95, R) >droSec1.super_0 18822873 110 + 21120651 AACAAUUA-----GUUUAAAUCGGUAAUUAAAAU-UGUGUCUAC-AAAGCGCAAGCGUAACUAUGAAUUAGAGAUGGAACACACUAAGGUGUUUACUCGAUACUAUCGAUAGUACAU------------------ ........-----((....(((((((........-.(((((...-...((....))..............(((...((((((......)))))).)))))))))))))))...))..------------------ ( -23.10, z-score = -1.58, R) >droYak2.chr3R 19314225 115 + 28832112 AGCAAUUUAUUGCGUUUAAAUCGGUAAUUAAAAUAUGUGUCUAC-AAUGCGCAAGCGUAACUAUGAAUUAGAGAUGGAACACACUAAGGUGUUUACUCGA-ACUAUCGAUCUUACAU------------------ .((((....))))((....(((((((..................-.((((....))))............(((...((((((......)))))).)))..-..)))))))...))..------------------ ( -24.80, z-score = -1.34, R) >droEre2.scaffold_4820 859583 115 - 10470090 UGCAAUUGAUUGCGCUUAAAUCCGUAAUUAAAAUAUGCGUCUAC-GAUGCGCAAGCGUAACUAUGAAUUAGAGAUGGAACACACUGAGGUGUU-ACGCGAUACUAUCAUGUGUUCAU------------------ .....(((((((((........)))))))))......(((((..-.((((....))))..(((.....))))))))(((((((.(((((((((-....)))))).))))))))))..------------------ ( -33.40, z-score = -2.37, R) >droAna3.scaffold_13340 21882730 131 + 23697760 GGUCAUUUAACUUGUUUAAAUCGACGAUAAAAAG--AUUUUACA-AACGCGCAAGCGUAACUACAAGUGUGUGACCGUACGCGG-AACGCGUAAAUAUACCACCUUACAAAAUAUACCAGAUUAUUGAGGUUCAU ((((((...((((((..(((((...........)--))))....-.((((....))))....))))))..)))))).((((((.-..))))))........((((((....(((........))))))))).... ( -34.30, z-score = -3.18, R) >dp4.chr2 26662731 112 - 30794189 UGCCAUUUAGUUGGUUAAAAACAA-AUUUGAAAG--ACGCCAA---ACGCGCAAGCGUAGC-GUAUAGUAGAGGUGUCAAACCAUCGAUGGCACUAUCGAUAGUAUCGAUGGUUUUUCU---------------- .((((((....(((((....(((.-.((((....--((((...---((((....)))).))-))....))))..)))..)))))..))))))(((((((((...)))))))))......---------------- ( -39.30, z-score = -4.91, R) >droPer1.super_7 223441 112 + 4445127 UGCCAUUUAGUUGGUUAAAAACAA-AUUUGAAAG--ACGCCUA---ACGCGCAAGCGUAGC-GUAUAGUAGAGGUGUCAAACCAUCGAUGGAACUAUCGAUAGUAUCGAUGGUUUUUCU---------------- .((((......)))).........-....(((((--((((((.---((((....)))).((-.....))..))))))).((((((((((...((((....)))))))))))))))))).---------------- ( -38.10, z-score = -4.60, R) >droWil1.scaffold_181089 8350371 100 - 12369635 -------UAUUAUUUUUUAAUAAA--ACUAAACCACAAAAC----AAAGCGCAAGCGCAGC--CUAGAUAAAAA--AAAACUAUUAAUGCAAACAGUUGGCAGCACCACUUUGGGUU------------------ -------(((((.....)))))..--....((((.((((..----...((....))((.((--(..........--......................))).)).....))))))))------------------ ( -15.95, z-score = -0.38, R) >droMoj3.scaffold_6540 5841734 105 - 34148556 -----UUGAUUAUAUUAAACCAAACAAUUAAAACAAAUCUCUUCAACUGCGCAAGCGUA-C--CCAGGUAAAAA--CACUUGGGUAUCACAAAAA--CAAAAGCAUCGAUACCAAAU------------------ -----(((((......................................((....))(((-(--((((((.....--.))))))))))........--.......)))))........------------------ ( -19.60, z-score = -3.84, R) >consensus AGCAAUUUAUUGUGUUUAAAUCGAUAAUUAAAAU_UAUGUCUAC_AAUGCGCAAGCGUAACUAUGAAUUAGAGAUGGAACACACUAAGGUGUAUACUCGAUACUAUCGAUAGUACAU__________________ ..............................................((((....))))......................((((....))))........................................... ( -5.24 = -5.37 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:48 2011