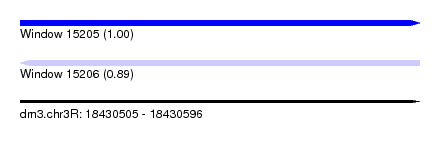
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,430,505 – 18,430,596 |
| Length | 91 |
| Max. P | 0.997256 |

| Location | 18,430,505 – 18,430,596 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.60201 |
| G+C content | 0.44766 |
| Mean single sequence MFE | -18.79 |
| Consensus MFE | -14.91 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
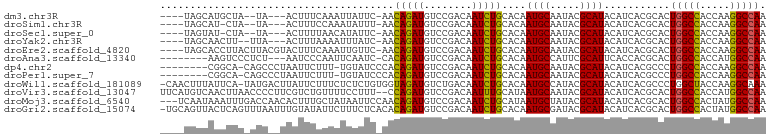
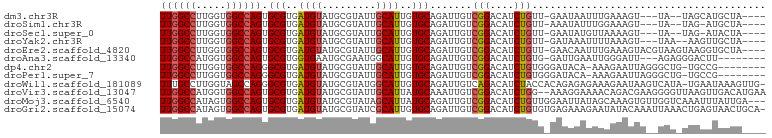
>dm3.chr3R 18430505 91 + 27905053 ----UAGCAUGCUA--UA---ACUUUCAAAUUAUUC-AACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAA ----..((......--..---...............-..(((((........)))))....((((.....)))).......))..(((((.....))))). ( -18.50, z-score = -1.94, R) >droSim1.chr3R 18241579 90 + 27517382 ----UAGCAU-CUA--UA---ACUUUCCAAAUAUUU-AACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAA ----..((..-...--..---...............-..(((((........)))))....((((.....)))).......))..(((((.....))))). ( -18.50, z-score = -2.79, R) >droSec1.super_0 18788678 90 + 21120651 ----UAGUAU-CUA--UA---ACUUUUAACAUAUUC-AACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAA ----..((..-...--..---...............-..(((((........)))))....((((.....)))).......))..(((((.....))))). ( -16.10, z-score = -1.86, R) >droYak2.chr3R 19279213 91 + 28832112 ----UAGCAACUU--UUA---ACUUUAAAAUUUAUC-AACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAA ----..((.....--...---...............-..(((((........)))))....((((.....)))).......))..(((((.....))))). ( -18.50, z-score = -2.91, R) >droEre2.scaffold_4820 824739 96 - 10470090 ----UAGCACCUUACUUACGUACUUUCAAAUUGUUC-AACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAA ----..((..................((.((((((.-.((....))..)))))).))....((((.....)))).......))..(((((.....))))). ( -18.70, z-score = -1.56, R) >droAna3.scaffold_13340 21846836 89 + 23697760 --------AAGUCCCUCU---AAUCCCAAUUCAAUC-CACAGAUGUCCGACAAUCUGCACAAUGCCAUUCGCAUUCACCACGCACUGGCCACCAUGGCCAA --------..........---...............-..(((((........)))))...(((((.....)))))..........(((((.....))))). ( -15.80, z-score = -1.39, R) >dp4.chr2 26619012 91 - 30794189 --------CGGCA-CAGCCCUAAUUCUUU-UGUAUCCCACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCCCUGGCCACCAAGGCCAA --------.(((.-...............-.........(((((........)))))....((((.....)))).......))).(((((.....))))). ( -21.00, z-score = -1.69, R) >droPer1.super_7 177660 91 + 4445127 --------CGGCA-CAGCCCUAAUUCUUU-UGUAUCCCACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCCCUGGCCACCAAGGCCAA --------.(((.-...............-.........(((((........)))))....((((.....)))).......))).(((((.....))))). ( -21.00, z-score = -1.69, R) >droWil1.scaffold_181089 8316782 99 - 12369635 -CAACUUUAUUCA-UAUGACUUAUUCUUUCUCUCUGUGGUAGAUGUCUGACAAUCUGCACAAUGCCAUACGCAUACAUCACGCCCUGGCUACCAAGGCAAA -............-.....................(((((((((........))))))...((((.....))))....)))(((.(((...))).)))... ( -19.10, z-score = -1.01, R) >droVir3.scaffold_13047 9024311 99 - 19223366 UUCAUGUCAACUUAACCCCUUCGUCUGUUUUCCUUU--CCAGAUGUCCGACAAUUUGCAUAAUGCAAUACGCAUACAUCACGCACUGGCCACCAUGGCCAA ....((((.............((((((.........--.))))))...))))..((((.....))))...((.........))..(((((.....))))). ( -17.99, z-score = -1.34, R) >droMoj3.scaffold_6540 5798953 98 - 34148556 ---UCAAUAAAUUUGACCAACACUUUGCUAUAAUUCCAACAGAUGUCCGACAAUCUGCAUAAUGCUAUACGCAUACAUCACGCACUGGCCACUAUGGCCAA ---((((.....)))).........(((...........(((((........)))))....((((.....)))).......))).(((((.....))))). ( -19.50, z-score = -2.35, R) >droGri2.scaffold_15074 3867226 100 + 7742996 -UGCAGUUACUCAGUUUAAUUUGUAUAUUCUUUCUCACACAGAUGUCCGACAAUCUGCACAAUGCGAUACGCAUACAUCACGCACUGGCCACUAUGGCCAA -(((((....((.(....((((((..............))))))..).))....)))))..(((((...)))))...........(((((.....))))). ( -20.74, z-score = -1.06, R) >consensus ____UAGCAAGCUA_AUA__UACUUUCAAAUUAUUC_AACAGAUGUCCGACAAUCUGCACAAUGCAAUACGCAUACAUCACGCACUGGCCACCAAGGCCAA .......................................(((((........)))))....((((.....))))...........(((((.....))))). (-14.91 = -14.77 + -0.14)
| Location | 18,430,505 – 18,430,596 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.60201 |
| G+C content | 0.44766 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -16.47 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.888320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
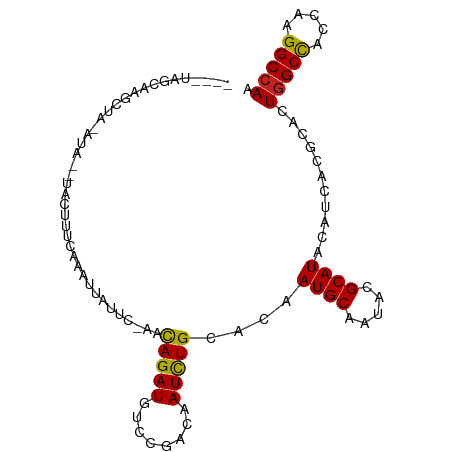
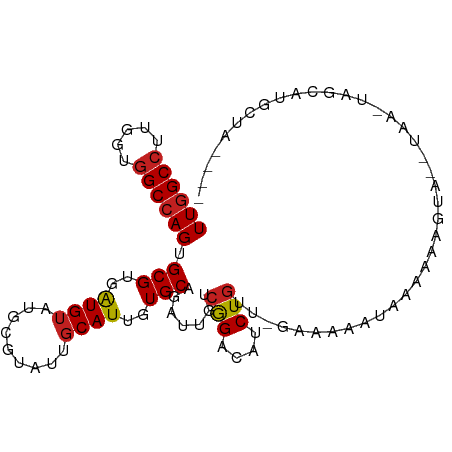
>dm3.chr3R 18430505 91 - 27905053 UUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUU-GAAUAAUUUGAAAGU---UA--UAGCAUGCUA---- ((((((.....))))))........((((((.(((.((.((...((((((((..(((....)))-..)))))))).))))---.)--))))))))..---- ( -26.40, z-score = -0.85, R) >droSim1.chr3R 18241579 90 - 27517382 UUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUU-AAAUAUUUGGAAAGU---UA--UAG-AUGCUA---- ((((((.....))))))((((..(((((.......)))))..)))).......((.(((((((.-...............---.)--)))-))))).---- ( -27.03, z-score = -1.59, R) >droSec1.super_0 18788678 90 - 21120651 UUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUU-GAAUAUGUUAAAAGU---UA--UAG-AUACUA---- ((((((.....))))))((((..(((((.......)))))..))))((((....(((((.(...-..).)))))...)))---).--...-......---- ( -23.70, z-score = -0.65, R) >droYak2.chr3R 19279213 91 - 28832112 UUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUU-GAUAAAUUUUAAAGU---UAA--AAGUUGCUA---- ((((((.....))))))((((..(((((.......)))))..))))..((((((.((....)))-))))).......(((---...--.....))).---- ( -24.90, z-score = -1.28, R) >droEre2.scaffold_4820 824739 96 + 10470090 UUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUU-GAACAAUUUGAAAGUACGUAAGUAAGGUGCUA---- ((((((.....)))))).(((..(((((.......)))))..)))(((((((..(((....)))-..)))))))...(((((.(.....).))))).---- ( -31.00, z-score = -1.89, R) >droAna3.scaffold_13340 21846836 89 - 23697760 UUGGCCAUGGUGGCCAGUGCGUGGUGAAUGCGAAUGGCAUUGUGCAGAUUGUCGGACAUCUGUG-GAUUGAAUUGGGAUU---AGAGGGACUU-------- .(((((.....)))))((.(.(....(((.(.(((..((.(.(((((((.(.....))))))))-.).)).))).).)))---..).).))..-------- ( -21.70, z-score = 0.63, R) >dp4.chr2 26619012 91 + 30794189 UUGGCCUUGGUGGCCAGGGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUGGGAUACA-AAAGAAUUAGGGCUG-UGCCG-------- ((((((.....)))))).(((..(((((.......)))))..))).......((((((.((.(((......-......))).)).))-).)))-------- ( -26.10, z-score = 0.33, R) >droPer1.super_7 177660 91 - 4445127 UUGGCCUUGGUGGCCAGGGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUGGGAUACA-AAAGAAUUAGGGCUG-UGCCG-------- ((((((.....)))))).(((..(((((.......)))))..))).......((((((.((.(((......-......))).)).))-).)))-------- ( -26.10, z-score = 0.33, R) >droWil1.scaffold_181089 8316782 99 + 12369635 UUUGCCUUGGUAGCCAGGGCGUGAUGUAUGCGUAUGGCAUUGUGCAGAUUGUCAGACAUCUACCACAGAGAGAAAGAAUAAGUCAUA-UGAAUAAAGUUG- ...(((((((...)))))))......(((.((((((((.(((((.((((.(.....)))))..)))))..(.......)..))))))-)).)))......- ( -21.80, z-score = 0.08, R) >droVir3.scaffold_13047 9024311 99 + 19223366 UUGGCCAUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUAUGCAAAUUGUCGGACAUCUGG--AAAGGAAAACAGACGAAGGGGUUAAGUUGACAUGAA ((((((.....))))))(((((((((((.......)))))))))))...(((((.((.(((..--..)))..(((...(....).)))..))))))).... ( -30.50, z-score = -2.38, R) >droMoj3.scaffold_6540 5798953 98 + 34148556 UUGGCCAUAGUGGCCAGUGCGUGAUGUAUGCGUAUAGCAUUAUGCAGAUUGUCGGACAUCUGUUGGAAUUAUAGCAAAGUGUUGGUCAAAUUUAUUGA--- ((((((.....))))))((((((((((.........))))))))))..(((.(.(((((.(((((......)))))..))))).).))).........--- ( -30.10, z-score = -1.98, R) >droGri2.scaffold_15074 3867226 100 - 7742996 UUGGCCAUAGUGGCCAGUGCGUGAUGUAUGCGUAUCGCAUUGUGCAGAUUGUCGGACAUCUGUGUGAGAAAGAAUAUACAAAUUAAACUGAGUAACUGCA- ..((((.....))))((((((..(((....)))..)))))).(((((.(((((((..((.((((((........)))))).))....)))).))))))))- ( -26.60, z-score = -0.44, R) >consensus UUGGCCUUGGUGGCCAGUGCGUGAUGUAUGCGUAUUGCAUUGUGCAGAUUGUCGGACAUCUGUU_GAAAAAUAAAAAAGUA__UAA_UAGCAUGCUA____ ((((((.....)))))).(((..((((.........))))..))).......(((....)))....................................... (-16.47 = -16.65 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:46 2011