
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,301,820 – 8,301,919 |
| Length | 99 |
| Max. P | 0.969157 |

| Location | 8,301,820 – 8,301,919 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 61.09 |
| Shannon entropy | 0.82722 |
| G+C content | 0.41832 |
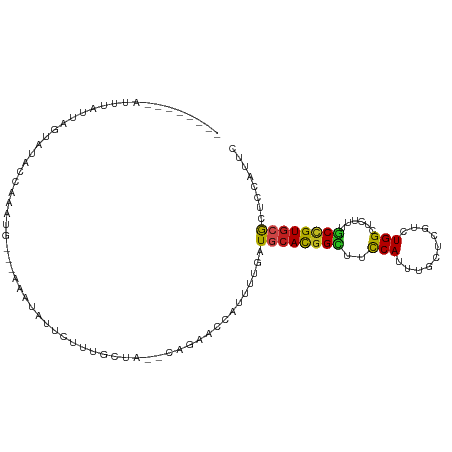
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -10.54 |
| Energy contribution | -10.05 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
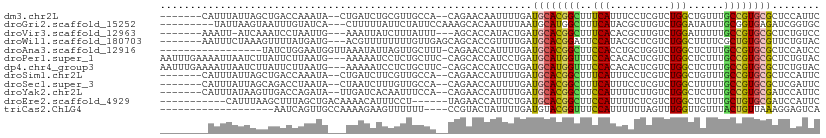
>dm3.chr2L 8301820 99 - 23011544 -------CAUUUAUUAGCUGACCAAAUA--CUGAUCUGCGUUGCCA--CAGAACAAUUUUGAUGCACGGCUUUCAUUUCCUCGUCUGGCUGUUUGCCGUGCGCUCCAUUC -------........(((....((((..--.((.((((.(....).--)))).))..))))..(((((((...((...((......)).))...))))))))))...... ( -24.40, z-score = -1.53, R) >droGri2.scaffold_15252 13492728 98 - 17193109 ---------UAUUAAGUAAUUUGUAUCA---CUUUUUAUUCUAUUCCAAAGCACAAUUUUAAUGCAUGGCUUUCAUACGCUUGUCUGGAUAUUUGCGGUGAGAUCGGUGC ---------((..((((.((....)).)---)))..)).........................((((.(.((((((.(((.(((....)))...))))))))).).)))) ( -14.50, z-score = 1.02, R) >droVir3.scaffold_12963 16056757 96 - 20206255 -------AAAUU-AUCAAAUCCUAAUUG---AAAUUAUCUUUAUUU---AGCACCAUACUGAUGCACGGCUUUCACACGCUUGUCUGGAUUUUUGCCGUGCGCUCUGUCC -------.....-........((((.((---((......)))).))---))......((.((((((((((.((((.((....)).)))).....)))))))).)).)).. ( -19.50, z-score = -1.81, R) >droWil1.scaffold_180703 3517057 100 + 3946847 -------AAUUUCUAAAGUUUUAUGAUG---ACGUUUUUUUUGUUGAGCAGCACCGUUUUGAUGCACGGAUUCCAUACGCUCGUCUGGCUUUUCGCUGUGCGUUCUGUAC -------...............(((.((---..((((........))))..)).)))...(((((((((...(((.((....)).))).......)))))))))...... ( -16.70, z-score = 1.56, R) >droAna3.scaffold_12916 10216842 92 + 16180835 -----------------UAUCUGGAAUGGUUAAAUAUUAGUUGCUUU-CAGAACCAUUUUGAUGCACGGCUUCCACCUGCUGGUCUGGCUCUUUGCCGUGCGCUCCAUCC -----------------.(((.(((((((((................-...))))))))))))(((((((..(((((....)))..))......)))))))......... ( -24.01, z-score = -1.04, R) >droPer1.super_1 4940950 106 + 10282868 AAUUUGAAAAUUAAUCUUAUUCUUAAUG---AAAAAUCCUCUGCUUC-CAGCACCAUCCUGAUGCAUGGUUUCCACACACUCGUCUGGCUCUUUGCCGUGCGCUCUGUAC ...................(((.....)---))........(((...-..))).......((((((((((..(((.((....)).)))......)))))))).))..... ( -16.20, z-score = -0.40, R) >dp4.chr4_group3 7830418 106 + 11692001 AAUUUGAAAAUUAAUCUUAUUCUUAAUG---AAAAAUCCUCUGCUUC-CAGCACCAUCCUGAUGCAUGGUUUCCACACACUCGUCUGGCUCUUUGCCGUGCGCUCUGUAC ...................(((.....)---))........(((...-..))).......((((((((((..(((.((....)).)))......)))))))).))..... ( -16.20, z-score = -0.40, R) >droSim1.chr2L 8089737 99 - 22036055 -------CAUUUAUUAGCUGACCAAAUA--CUGAUCUUCGUUGCCA--CAGAACCAUUUUGAUGCACGGCUUUCAUUUCCUCGUCUGGCUGUUUGCCGUGCGCUCCAUUC -------........(((..........--................--((((.....))))..(((((((...((...((......)).))...))))))))))...... ( -19.00, z-score = -0.28, R) >droSec1.super_3 3797974 99 - 7220098 -------CAUUUAUUAGCAGACCUAAUA--CUAAUCUUUGUUGCCA--CAGAACCAUUUUGAUGCACGGCUUUCAUUUCCUCGUCUGGCUUUUUGCCGUGCGCUCGAUUC -------....((((((.....))))))--......(((((....)--))))......((((((((((((........((......))......)))))))).))))... ( -21.04, z-score = -1.50, R) >droYak2.chr2L 10939625 99 - 22324452 -------CAUUUAUAAGUUGACCAGAUA--UUGAUCACAAUUUCCA--CAGAACCAUUUUGAUGCACGGCUUCCAUUUUCUUGUCUGGCUCUUUGCCGUGCGAUCCAUUC -------.....................--..((((..........--((((.....))))..(((((((..(((..........)))......)))))))))))..... ( -18.50, z-score = -0.68, R) >droEre2.scaffold_4929 17211316 93 - 26641161 -----------CAUUUAAGCUUUAGCUGACAAAACAUUUCCU------UAGAACCAUUCUGAUGCACGGCUUCCAUUUUCUCGUCUGGCUCUUUGCUGUGCGAUCCAUUC -----------......(((....)))..............(------((((.....)))))((((((((..(((..........)))......))))))))........ ( -16.00, z-score = -0.17, R) >triCas2.ChLG4 12475763 87 + 13894384 -------------------AAUCAGUUGCCAAAAGAAGUUUUUU----CCGUACUAUUUUGAUGUACGGUUUCCAUUUUUUAGUUUGGUUGUUUACUGUUAAAGGAGUCA -------------------...((((.((((((((((((.....----((((((.........)))))).....))))))...)))))).....))))............ ( -17.90, z-score = -1.92, R) >consensus ________AUUUAUUAGUAUACCAAAUG___AAAUAUUCUUUGCUA__CAGAACCAUUUUGAUGCACGGCUUCCAUUUGCUCGUCUGGCUCUUUGCCGUGCGCUCCAUUC ..............................................................((((((((..(((..........)))......))))))))........ (-10.54 = -10.05 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:42 2011