| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,425,867 – 18,425,960 |
| Length | 93 |
| Max. P | 0.976406 |

| Location | 18,425,867 – 18,425,960 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.98 |
| Shannon entropy | 0.34911 |
| G+C content | 0.62835 |
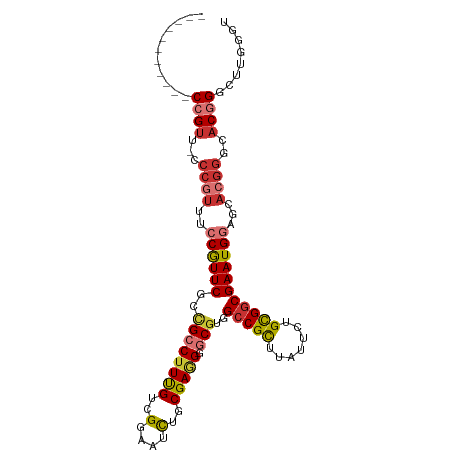
| Mean single sequence MFE | -46.84 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.79 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976406 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
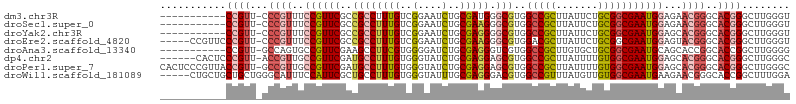
>dm3.chr3R 18425867 93 - 27905053 -----------CCGUU-CCCGUUUCCGUUCGCCGCCUUUGUCGGAAUCUGCGAUGGGCGUGGCCGCUUAUUCUGCGGCGAAUGGAGAACGGGCACGGGCUUGGGU -----------((((.-(((((((((((((.((((((..((((.......))))))))).)(((((.......))))))))))))).))))).))))........ ( -47.00, z-score = -2.95, R) >droSec1.super_0 18784060 93 - 21120651 -----------CCGUU-CCCGUUUCCGUUCGCCGCCUUUGUCGGAAUCUGCGAAGGGCGUGGCCGCUUAUUCUGCGGCGAAUGGAGAACGGGCACGGGCUUGGGU -----------((((.-(((((((((((((((((((((((.(((...))))))))((((....))))......))))))))))))).))))).))))........ ( -48.30, z-score = -3.24, R) >droYak2.chr3R 19274593 93 - 28832112 -----------CCGUU-CCCGUUUCCGUUCGCCGCCUUUGUCGGAAUCUGCGAGGGGCGUGGCCGCUUAUUCUGCGGCGAAUGGAGCACGGGCACGGGCUUGGGU -----------((((.-(((((((((((((((((((((((.(((...))))))))((((....))))......))))))))))))).))))).))))........ ( -48.00, z-score = -2.67, R) >droEre2.scaffold_4820 820159 99 + 10470090 -----CCGUUCCCGUU-CCCGUUUCCGUUCGCCGCCUUUGUCGGAAUCUGCGAAGGGCGUGGACGCUUAUUCUGCGGCGAAUGGAGUACGGGCACGGGCUUGGGU -----(((..(((((.-(((((((((((((((((((((((.(((...))))))))((((....))))......))))))))))))).))))).)))))..))).. ( -56.40, z-score = -5.10, R) >droAna3.scaffold_13340 21842070 93 - 23697760 -----------CCGUU-GCCAGUGCCGUUCGAAGCCUUCGUGGGGAUCUGCGAGGGUCGUGGCCGCUUGUGCUGCGGCGAAUGCAGCACCGGCACCGGCUUGGGG -----------(((..-(((.((((((..((..(((((((..(....)..)))))))..))..)....((((((((.....)))))))).))))).))).))).. ( -48.60, z-score = -2.11, R) >dp4.chr2 26611416 98 + 30794189 ------CACUCCCGUU-ACCGUUGCCGUUCGAUGCCUUUGUGGGUAUCUGCGAGGAGCGUGGCCGCUUAUUUUGUGGCGAAUGGAGCACGGGCACGGGCUUGGGC ------((..(((((.-.((((..((((((.(((((((((..(....)..))))).)))).(((((.......)))))))))))...))))..)))))..))... ( -44.80, z-score = -2.52, R) >droPer1.super_7 172341 104 - 4445127 CACUCCCGUUACCGUU-GCCGUUGCCGUUCGAUGCCUUUGUGGGUAUCUGCGAGGAGCGUGGCCGCUUAUUUUGUGGCGAAUGGAGCACGGGCACGGGCUUGGGC ....((((...((((.-(((((..((((((.(((((((((..(....)..))))).)))).(((((.......)))))))))))...)).)))))))...)))). ( -46.30, z-score = -1.95, R) >droWil1.scaffold_181089 8311900 100 + 12369635 -----CUGCUGCUGCUGGGCAUUUCCAUUCGCUGCCUUUGUGGGUAUUUGCGAGGGACGUGGCCGUUUAUGUUGUGGCGAAUGAAGAACGGGCACCGGCUUUGGA -----..((((.((((.(...(((.(((((((..((((((..(....)..)))))(((((((....))))))))..))))))).))).).)))).))))...... ( -35.30, z-score = -0.72, R) >consensus ___________CCGUU_CCCGUUUCCGUUCGCCGCCUUUGUCGGAAUCUGCGAGGGGCGUGGCCGCUUAUUCUGCGGCGAAUGGAGCACGGGCACGGGCUUGGGU ...........((((...((((..((((((..((((((((..(....)..))))).)))..(((((.......)))))))))))...))))..))))........ (-27.92 = -28.79 + 0.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:43 2011