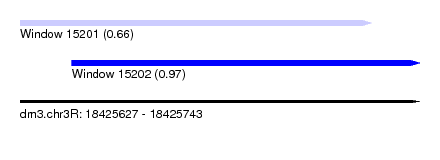
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,425,627 – 18,425,743 |
| Length | 116 |
| Max. P | 0.971445 |

| Location | 18,425,627 – 18,425,729 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.31 |
| Shannon entropy | 0.54443 |
| G+C content | 0.57096 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -11.32 |
| Energy contribution | -12.58 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
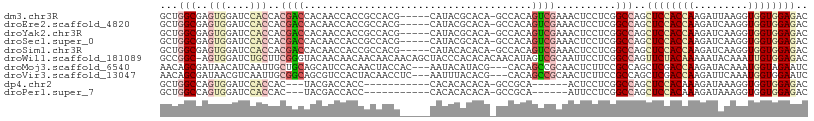
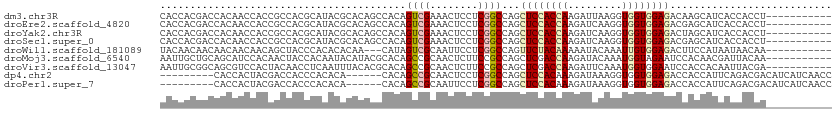
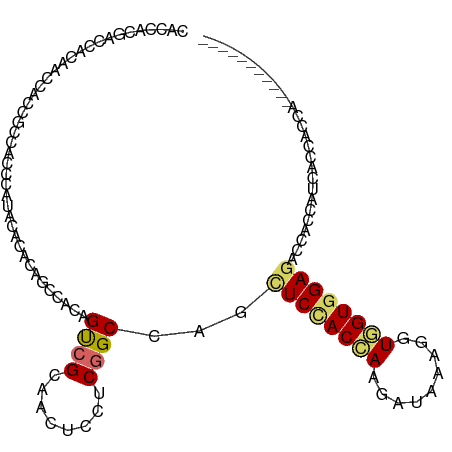
>dm3.chr3R 18425627 102 + 27905053 GCUGGCGAGUGGAUCCACCACGACCACAACCACCGCCACG-----CAUACGCACA-GCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUUAAGGUGGUGGAGAC (((((((.((((.((......)))))).......((...)-----)...))).))-))....(((((......)))))...((((((((.........)))))))).. ( -33.80, z-score = -2.23, R) >droEre2.scaffold_4820 819922 102 - 10470090 GCUGGCGAGUGGAUCCACCACGACCACAACCACCGCCACG-----CAUACGCACA-GCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUCAAGGUGGUGGAGAC (((((((.((((.((......)))))).......((...)-----)...))).))-))....(((((......)))))...((((((((.........)))))))).. ( -33.80, z-score = -1.98, R) >droYak2.chr3R 19274353 102 + 28832112 GCUGGCGAGUGGAUCCACCACGACCACAACCACCGCCACG-----CAUACGCACA-GCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUCAAGGUGGUGGAGAC (((((((.((((.((......)))))).......((...)-----)...))).))-))....(((((......)))))...((((((((.........)))))))).. ( -33.80, z-score = -1.98, R) >droSec1.super_0 18783832 102 + 21120651 GCUGGCGAGUGGAUCCACCACGACCACAACCACCGCCACG-----CAUACGCACA-GCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUCAAGGUGGUGGAGAC (((((((.((((.((......)))))).......((...)-----)...))).))-))....(((((......)))))...((((((((.........)))))))).. ( -33.80, z-score = -1.98, R) >droSim1.chr3R 18237169 102 + 27517382 GCUGGCGAGUGGAUCCACCACGACCACAACCACCGCCACG-----CAUACACACA-GCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUCAAGGUGGUGGAGAC (((((((.((((.................))))))))).)-----).........-......(((((......)))))...((((((((.........)))))))).. ( -33.33, z-score = -2.23, R) >droWil1.scaffold_181089 8311661 107 - 12369635 GCCGGC-AGUGGAUCUGCUUCGGGUACAACAACAACAACAACAGCUACCCACACACAACAUAGUCGCAAUUCCUCGGCCAGUUCUACAAAAAUACAAAUUGUGGAGAC ...(((-.(.(((..(((...(((((...................)))))....((......)).)))..))).).)))..((((((((.........)))))))).. ( -24.21, z-score = -1.51, R) >droMoj3.scaffold_6540 5794147 102 - 34148556 AACAGCGAUAACAUCAAUUGCUGCAGCAUCCACAACUACCAC---AAUACAUACG---CACAGCCGCAACUCUUCCGCCAGCUCGACCAAGAUACAAAUGGUAGAAUC ..(((((((.......)))))))............((((((.---.........(---(......))...((((.((......))...))))......)))))).... ( -16.40, z-score = -1.16, R) >droVir3.scaffold_13047 9019558 102 - 19223366 AACAGCGAUAACGUCAAUUGCGGCAGCGUCCACUACAACCUC---AAUUUACACG---CACAGCCGCAACUCUUCCGCCAGCUCGACCAAGAUUCAAAUGGUGGAAUC ....(((....)))...(((((((.((((.............---.......)))---)...)))))))...((((((((..((......))......)))))))).. ( -24.95, z-score = -1.85, R) >dp4.chr2 26611206 87 - 30794189 GCUGGCCAGUGGAUCCACCAC---UACGACCACC-----------CACACACACA-GCCGCA------ACUCCUCGGCCAGCUCCACAAAGAUAAAGGUGGUGGAGAC ((((((((((((.....))))---)..(......-----------).........-......------.......)))))))(((((.............)))))... ( -25.02, z-score = -0.96, R) >droPer1.super_7 172131 87 + 4445127 GCUGGCCAGUGGAUCCACCAC---UACGACCACC-----------CACACACACA-GCCGCA------AUUCCUCGGCCAGCUCCACAAAGAUAAAGGUGGUGGAGAC ((((((((((((.....))))---)..(......-----------).........-......------.......)))))))(((((.............)))))... ( -25.02, z-score = -1.12, R) >consensus GCUGGCGAGUGGAUCCACCACGACCACAACCACCGCCACG_____CAUACACACA_GCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUAAAGGUGGUGGAGAC ...(((..(((....)))..((((......................................))))..........)))..((((((((.........)))))))).. (-11.32 = -12.58 + 1.26)
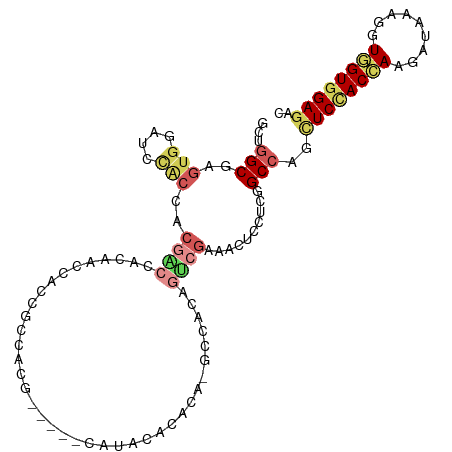
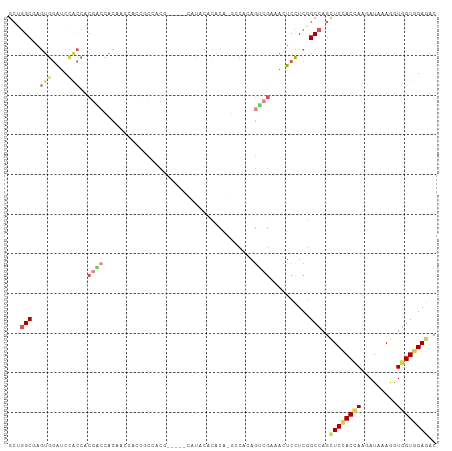
| Location | 18,425,642 – 18,425,743 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.38 |
| Shannon entropy | 0.63548 |
| G+C content | 0.53859 |
| Mean single sequence MFE | -19.61 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.60 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18425642 101 + 27905053 CACCACGACCACAACCACCGCCACGCAUACGCACAGCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUUAAGGUGGUGGAGACAAGCAUCACCACCU----------- ..............((((((((..((....))..(((....(((((......)))))..))).............))))))))..................----------- ( -24.60, z-score = -2.16, R) >droEre2.scaffold_4820 819937 101 - 10470090 CACCACGACCACAACCACCGCCACGCAUACGCACAGCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUCAAGGUGGUGGAGACGAGCAUCACCACCU----------- ..............((((((((..((....))..(((....(((((......)))))..))).............))))))))..................----------- ( -24.60, z-score = -1.58, R) >droYak2.chr3R 19274368 101 + 28832112 CACCACGACCACAACCACCGCCACGCAUACGCACAGCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUCAAGGUGGUGGAGACUAGCAUCACCACCU----------- ..............((((((((..((....))..(((....(((((......)))))..))).............))))))))..................----------- ( -24.60, z-score = -1.91, R) >droSec1.super_0 18783847 101 + 21120651 CACCACGACCACAACCACCGCCACGCAUACGCACAGCCACAGUCGAAACUCCUCGGCCAGCUCCACCAAGAUCAAGGUGGUGGAGACGAGCAUCACCACCU----------- ..............((((((((..((....))..(((....(((((......)))))..))).............))))))))..................----------- ( -24.60, z-score = -1.58, R) >droWil1.scaffold_181089 8311684 98 - 12369635 UACAACAACAACAACAACAGCUACCCACACACAA---CAUAGUCGCAAUUCCUCGGCCAGUUCUACAAAAAUACAAAUUGUGGAGACUUCCAUAAUAACAA----------- ..................................---....((((........))))...................(((((((......))))))).....----------- ( -9.30, z-score = -0.48, R) >droMoj3.scaffold_6540 5794162 101 - 34148556 AAUUGCUGCAGCAUCCACAACUACCACAAUACAUACGCACAGCCGCAACUCUUCCGCCAGCUCGACCAAGAUACAAAUGGUAGAAUCCACAACGAUUACAA----------- ...(((....))).......((((((..........((......))...((((.((......))...))))......))))))((((......))))....----------- ( -11.90, z-score = 0.02, R) >droVir3.scaffold_13047 9019573 101 - 19223366 AAUUGCGGCAGCGUCCACUACAACCUCAAUUUACACGCACAGCCGCAACUCUUCCGCCAGCUCGACCAAGAUUCAAAUGGUGGAAUCCACCACAAUUACGA----------- ..(((((((.((((....................))))...)))))))...((((((((..((......))......))))))))................----------- ( -24.65, z-score = -2.75, R) >dp4.chr2 26611221 97 - 30794189 ---------CACCACUACGACCACCCACACA------CACAGCCGCAACUCCUCGGCCAGCUCCACAAAGAUAAAGGUGGUGGAGACCACCAUUCAGACGACAUCAUCAACC ---------........((............------....((((........))))..................((((((....)))))).......))............ ( -16.10, z-score = -0.41, R) >droPer1.super_7 172146 97 + 4445127 ---------CACCACUACGACCACCCACACA------CACAGCCGCAAUUCCUCGGCCAGCUCCACAAAGAUAAAGGUGGUGGAGACCACCAUUCAGACGACAUCAUCAACC ---------........((............------....((((........))))..................((((((....)))))).......))............ ( -16.10, z-score = -0.52, R) >consensus CACCACGACCACAACCACCGCCACCCAUACACACAGCCACAGUCGCAACUCCUCGGCCAGCUCCACCAAGAUAAAGGUGGUGGAGACCACCAUCACCACCA___________ .........................................((((........))))...((((((((.........))))))))........................... (-11.25 = -11.60 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:42 2011