| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,422,107 – 18,422,209 |
| Length | 102 |
| Max. P | 0.840604 |

| Location | 18,422,107 – 18,422,209 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Shannon entropy | 0.38478 |
| G+C content | 0.51099 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -18.81 |
| Energy contribution | -20.00 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
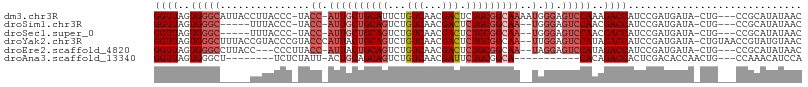
>dm3.chr3R 18422107 102 + 27905053 GGUUAGUGGGCAUUACCUUACCC-UACC-AUUGCUGCAUUCUGUCAACGACUCUGCGGCAAAAUGGGAGUCCAAAGACCAUCCGAUGAUA-CUG---CCGCAUAUAAC (((.((((..((((......(((-....-.((((((((....(((...)))..))))))))...))).(((....))).....)))).))-)))---))......... ( -26.40, z-score = -0.91, R) >droSim1.chr3R 18233632 95 + 27517382 GGUUAGUGGGC-----UUUACCC-UACC-AUUGCUGCAGUCUGUCAACGACUCUGCGGCAA--UGGGAGUCCAACGACCAUCCGAUGAUA-CUG---CCGCAUAUAAC ((((..(((((-----((.....-..((-((((((((((...(((...))).)))))))))--))))))))))..))))...........-...---........... ( -36.31, z-score = -3.73, R) >droSec1.super_0 18774777 95 + 21120651 GGUUAGUGGGC-----UUUACCC-UACC-AUUGCUGCAGUCUGUCAACGACUCUGCGGCAA--UGGGAGUCCAACGACCAUCCGAUGAUA-CUG---CCGCAUAUAAC ((((..(((((-----((.....-..((-((((((((((...(((...))).)))))))))--))))))))))..))))...........-...---........... ( -36.31, z-score = -3.73, R) >droYak2.chr3R 19270738 105 + 28832112 GGUUAGUGGGCUUUACCGUACCCGUACCCAUUACUGCAGUCUGUCAACGACUCUGCGGCAA--UUGGAGUCCAUAGACCAUCCGAUGAUA-CUGUAACCGUAUGUAAC ((((.(((((((.....(((....)))(((...((((((...(((...))).))))))...--.)))))))))).))))........(((-(.......))))..... ( -29.10, z-score = -0.80, R) >droEre2.scaffold_4820 816464 98 - 10470090 GGUUAGUGGGCCUUACC---CCCUUACC-AUUACUGCAGUCUGUCAACGACUCUGCGGCAA--UAGGAGUCCAUAGACCAUCCGAUGAUA-CUG---CCGCAUAUAAC (((.((.(((.....))---).)).)))-........((((.......)))).((((((((--(.((((((....)))..)))....)).-.))---)))))...... ( -26.00, z-score = -0.93, R) >droAna3.scaffold_13340 21838077 85 + 23697760 GGUUAGUGGGCU--------UCUCUAUU-ACUGCAGCAGUCUGUCAACGAUUCUGCGGCA-----------CACAGACCACUCGACACCAACUG---CCAAACAUCCA (((.(((((...--------(((.....-.((((((.((((.......))))))))))..-----------...))))))))....))).....---........... ( -18.80, z-score = -0.17, R) >consensus GGUUAGUGGGC_____CUUACCC_UACC_AUUGCUGCAGUCUGUCAACGACUCUGCGGCAA__UGGGAGUCCAAAGACCAUCCGAUGAUA_CUG___CCGCAUAUAAC ((((..(((((...................(((((((((...(((...))).))))))))).......)))))..))))............................. (-18.81 = -20.00 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:41 2011