| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,418,918 – 18,419,014 |
| Length | 96 |
| Max. P | 0.922610 |

| Location | 18,418,918 – 18,419,014 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Shannon entropy | 0.41224 |
| G+C content | 0.48095 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -18.15 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
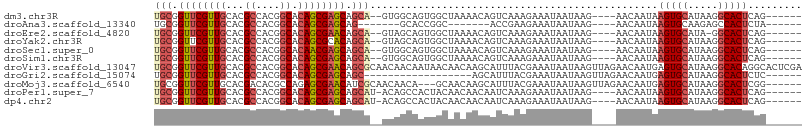
>dm3.chr3R 18418918 96 + 27905053 UGCGGUUCGUUGCACGCCACGGCACAGCGAGCAGCA--GUGGCAGUGGCUAAAACAGUCAAAGAAAUAAUAAG----AACAAUAAGUGCAUAAGGCACUCAG------ (((.((((((((...((....)).)))))))).)))--((..(..(((((.....)))))..)..))......----.......(((((.....)))))...------ ( -26.30, z-score = -1.27, R) >droAna3.scaffold_13340 21835222 84 + 23697760 UGCGGUUCGUUGCACGCCACGGCACAGCGAGCAG-------GCACCGGC-------ACCGAAGAAAUAAUAAG----AACAAUAAGUGCAAGAGCCACUCUA------ ...(((((.(((((((((...((.......)).)-------))..((..-------..)).............----........)))))))))))......------ ( -24.30, z-score = -1.30, R) >droEre2.scaffold_4820 813277 95 - 10470090 UGCGGUUCGUUGCACGCCACGGCACAGCGAACAGCA--GUAGCAGUGGCUAAAACAGUCAAAGAAAUAAUAAG----AACAAUAAGUGCAUA-GGCACUCAG------ (((.((((((((...((....)).)))))))).)))--.......(((((.....))))).............----.......(((((...-.)))))...------ ( -25.40, z-score = -1.83, R) >droYak2.chr3R 19267530 96 + 28832112 UGCGGUUCGUUGCACGCCACGGCACAGCGCACAGCA--GUAGCAGUGGCUAAAACAGUCAAAGAAAUAAUAAG----AACAAUAAGUGCAUAAGGCACUCAG------ ....(((((((....(((((.((((.((.....)).--)).)).)))))...)))......(....).....)----)))....(((((.....)))))...------ ( -24.20, z-score = -1.00, R) >droSec1.super_0 18771721 96 + 21120651 UGCGGUUCGUUGCACGCCACGGCACAACGAGCAGCA--GUGGCAGUGGCUAAAACAGUCAAAGAAAUAAUAAG----AACAAUAAGUGCAUAAGGCACUCAG------ (((.((((((((...((....)).)))))))).)))--((..(..(((((.....)))))..)..))......----.......(((((.....)))))...------ ( -26.30, z-score = -1.67, R) >droSim1.chr3R 18230556 96 + 27517382 UGCGGUUCGUUGCACGCCACGGCACAGCGAGCAGCA--GUGGCAGUGGCUAAAACAGUCAAAGAAAUAAUAAG----AACAAUAAGUGCAUAAGGCACUCAG------ (((.((((((((...((....)).)))))))).)))--((..(..(((((.....)))))..)..))......----.......(((((.....)))))...------ ( -26.30, z-score = -1.27, R) >droVir3.scaffold_13047 9014449 108 - 19223366 UGCGGUUCGUUGCACGCCACGGCACAGCGAACAGCGCAACAACAAUAACAACAAGCAUUUACGAAAUAAUAAGUUAGAACAAUGAGUGCAUAAGGCACAGGCACUCGA ((((((((((((...((....)).))))))))..))))............................................(((((((...........))))))). ( -25.50, z-score = -1.19, R) >droGri2.scaffold_15074 3856145 84 + 7742996 UGCGGUUCGUUGCACGCCACGGCACAGCGAGCAGC------------------AGCAUUUACGAAAUAAUAAGUUAGAACAAUGAGUGCAUAAGGCACUCUC------ (((.((((((((...((....)).)))))))).))------------------).............................((((((.....))))))..------ ( -25.70, z-score = -2.14, R) >droMoj3.scaffold_6540 5789184 99 - 34148556 UGCGGUUCGUUGCACGACACGCCAGAGCGAACAUCGCAACAACA---GCAACAAGCAUUUACGAAAUAAUAAGUUAGAACAAUGAGUGCAUAAGGCACUCGG------ ....(((((((((......(((....)))......)))))....---((.....))....................))))..(((((((.....))))))).------ ( -24.80, z-score = -1.79, R) >droPer1.super_7 166646 97 + 4445127 UGCGGUUCGUUGCACGCCACGGCACAGCGAGCAGCAU-ACAGCCACUACAACAACAAUCAAAGAAAUAAUAAG----AACAAUAAGUGCAUAAGGCACUCAG------ (((.((((((((...((....)).)))))))).))).-...................................----.......(((((.....)))))...------ ( -21.80, z-score = -1.83, R) >dp4.chr2 26605661 97 - 30794189 UGCGGUUCGUUGCACGCCACGGCACAGCGAGCAGCAU-ACAGCCACUACAACAACAAUCAAAGAAAUAAUAAG----AACAAUAAGUGCAUAAGGCACUCAG------ (((.((((((((...((....)).)))))))).))).-...................................----.......(((((.....)))))...------ ( -21.80, z-score = -1.83, R) >consensus UGCGGUUCGUUGCACGCCACGGCACAGCGAGCAGCA__GUAGCAGUGGCUAAAACAAUCAAAGAAAUAAUAAG____AACAAUAAGUGCAUAAGGCACUCAG______ .((.((((((((...((....)).)))))))).)).................................................(((((.....)))))......... (-18.15 = -18.47 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:38 2011