| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,401,065 – 18,401,170 |
| Length | 105 |
| Max. P | 0.989575 |

| Location | 18,401,065 – 18,401,170 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Shannon entropy | 0.43430 |
| G+C content | 0.48627 |
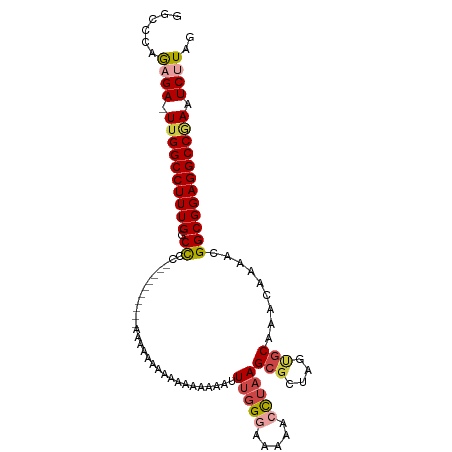
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989575 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
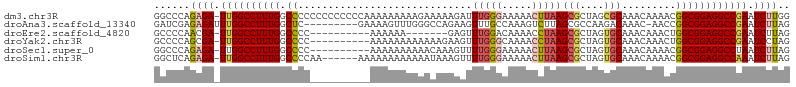
>dm3.chr3R 18401065 105 - 27905053 GGCCCAGAGA-UUGGCCUUUGGCCCCCCCCCCCCCAAAAAAAAAGAAAAAGAUUUGGGAAAAACUUAAGCGCUAGCGCAAACAAAACGGCGGAGGCCGAAUCUUGG ((((......-..))))(((((((.((((...((((((..............))))))..........(((....))).........)).)).)))))))...... ( -32.34, z-score = -1.08, R) >droAna3.scaffold_13340 21816348 96 - 23697760 GAUCGAGAGAUUUGGCCUUUGGCUC---------GAAAAGUUUGGGCCAGAAGUUUGCCAAAGUCUUAGCGCCAAGACAAAC-AACCGGCGGAGGCCGAAUCUUAG ......(((((((((((((((((((---------((.....))))))))))..((((((...(((((......)))))....-....))))))))))))))))).. ( -37.70, z-score = -3.45, R) >droEre2.scaffold_4820 795405 88 + 10470090 GCCCCAACGA-UUGGCCUUUGGCCCC----------AAAAAA-------GAGUUUGGACAAAACCUAAGCGCUAGUGCAAACAAACUGGCGGAGGCCGAAUCUUAG (((.......-..))).(((((((((----------......-------..((((((.......))))))((((((........)))))))).)))))))...... ( -29.10, z-score = -2.42, R) >droYak2.chr3R 19249554 95 - 28832112 GCCCCAGCGA-UUGGCCUUUGGCCCC----------AAAAAAAAAAAAGAAGUUUGGGCAAAACCUAAGCGCUAGUGCAAACAAACUGGCGGAGGCCGAAUCCUAG ((....))..-.(((..(((((((((----------...............(((((((.....)))))))((((((........)))))))).)))))))..))). ( -32.70, z-score = -2.36, R) >droSec1.super_0 18754163 95 - 21120651 GGCCCAGAGA-UUGGCCUUUGGCCCC----------AAAAAAAAAACAAAGUUUUGGGAAAAACUUAAGCGCUAGUGCAAACAAAACGGCGGAGGCCUAAUCUUAG ......((((-((((((((((.((((----------((((...........)))))))..........(((....))).........).)))))))).)))))).. ( -29.60, z-score = -1.99, R) >droSim1.chr3R 18212605 99 - 27517382 GGCUCAGAGA-UUGGCCUUUGGCCCCAA------AAAAAAAAAAAAUAAAGUUUUGGGAAAAACUUAAGCGCUAGUGCAAACAAAACGGCGGAGGCCAAAUCUUAG ......((((-((((((((((.((((((------((...............)))))))..........(((....))).........).)))))))).)))))).. ( -29.26, z-score = -2.44, R) >consensus GGCCCAGAGA_UUGGCCUUUGGCCCC__________AAAAAAAAAAAAAAAAUUUGGGAAAAACCUAAGCGCUAGUGCAAACAAAACGGCGGAGGCCGAAUCUUAG ......((((.((((((((((.((.............................(((((.....)))))(((....))).........)))))))))))).)))).. (-20.85 = -21.60 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:37 2011