
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,379,858 – 18,379,923 |
| Length | 65 |
| Max. P | 0.520687 |

| Location | 18,379,858 – 18,379,923 |
|---|---|
| Length | 65 |
| Sequences | 11 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 90.51 |
| Shannon entropy | 0.19952 |
| G+C content | 0.38281 |
| Mean single sequence MFE | -13.74 |
| Consensus MFE | -8.72 |
| Energy contribution | -8.81 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18379858 65 - 27905053 UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUUA-AUACUAUCU-UUGGCAUUCAUCUUG ((((((((((....)))......)))))))..((((((.((-....)).))-))))........... ( -11.00, z-score = -1.61, R) >droSim1.chr3R 18189569 65 - 27517382 UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUUA-AUACUAACU-UUGGCAUUCAUCUUG ((((((((((....)))......)))))))..(((((((((-....)))))-))))........... ( -15.10, z-score = -3.52, R) >droSec1.super_0 18733010 65 - 21120651 UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUUA-AUACUAACU-UUGGCAUUCAUCUUG ((((((((((....)))......)))))))..(((((((((-....)))))-))))........... ( -15.10, z-score = -3.52, R) >droYak2.chr3R 19227442 67 - 28832112 UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUUAAAGACUAACUCUUGGCAUUCAUCUUG .(((((((((....)))......))))))((.(...(((((.....)))))...))).......... ( -11.80, z-score = -1.17, R) >droEre2.scaffold_4820 773008 65 + 10470090 UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUUA-AGACUAACU-UUGGCAUUCAUCUCG ((((((((((....)))......)))))))..(((((((((-....)))))-))))........... ( -15.10, z-score = -2.84, R) >droAna3.scaffold_13340 21794973 64 - 23697760 UCAAGGCUUAAUUUGAAUUUUUCGCCUUGGCACUAAAGUU--AGUAUAACU-UUGGCAUUCAUCUUG (((((((..((........))..)))))))..((((((((--(...)))))-))))........... ( -14.60, z-score = -2.61, R) >dp4.chr2 27495298 64 + 30794189 UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUC--GAUCUAACU-UUGGCAUUCAUCUUG ((((((((((....)))......)))))))..(((((((.--......)))-))))........... ( -12.30, z-score = -1.64, R) >droPer1.super_7 1042751 64 - 4445127 UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUC--GAUCUAACU-UUGGCAUUCAUCUUG ((((((((((....)))......)))))))..(((((((.--......)))-))))........... ( -12.30, z-score = -1.64, R) >droWil1.scaffold_181130 1416140 64 + 16660200 UCAAGGCUUUAUUUGAAUUUCUUGCCUUGGCACUAAAGUUA--GACCAACU-UUGGCAUUCAUCUUG (((((((................)))))))..((((((((.--....))))-))))........... ( -13.99, z-score = -1.76, R) >droMoj3.scaffold_6540 24901421 62 + 34148556 -CAAGGCUAGAUUUGAAUUUUUCGCCUUGGCACUAAAGUC---GUAUAACU-UUGGCAUUCAUCUUG -((((((.(((........))).))))))...(((((((.---.....)))-))))........... ( -14.30, z-score = -2.16, R) >droGri2.scaffold_14906 6815778 63 + 14172833 UCAAGGCUGGAUUUGAAUUAUUCGCCUUGGCACUAAAGUC---GUAUAACU-UUGGCAUUCAUCUUG (((((((.((((.......)))))))))))..(((((((.---.....)))-))))........... ( -15.60, z-score = -2.33, R) >consensus UCAAGGCUUCAUUUGAAUUUCUCGCCUUGGCACUAAAGUUA_AGACUAACU_UUGGCAUUCAUCUUG .((((((................))))))((.(...(((.........)))...))).......... ( -8.72 = -8.81 + 0.09)

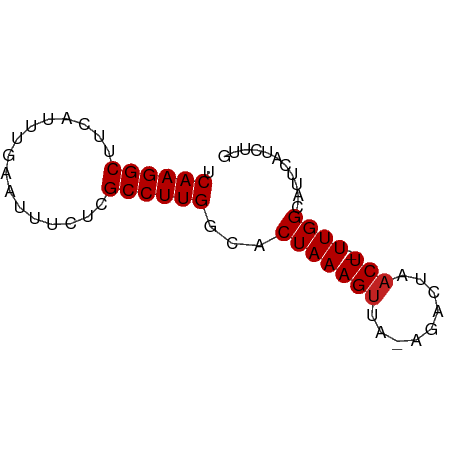
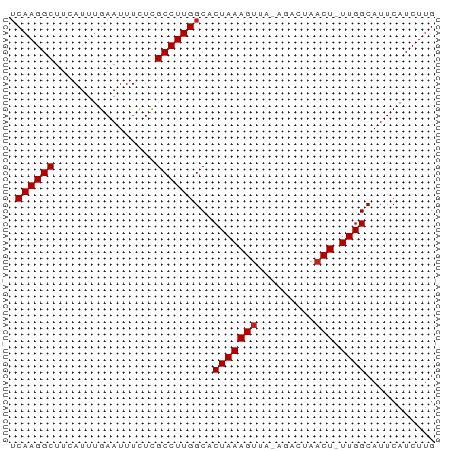
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:33 2011