| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,376,901 – 18,377,007 |
| Length | 106 |
| Max. P | 0.896990 |

| Location | 18,376,901 – 18,377,007 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.77 |
| Shannon entropy | 0.72862 |
| G+C content | 0.47853 |
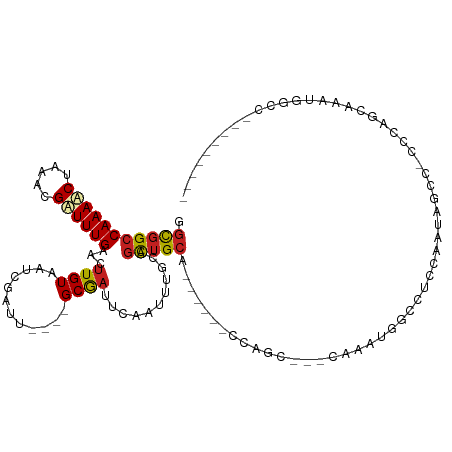
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -9.46 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
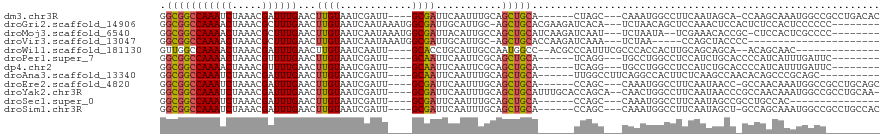
>dm3.chr3R 18376901 106 + 27905053 GGCGGCCAAAUCUAAACGAUUUGAACUUGUAAUCGAUU----GCGAUUCAAUUUGCAGCUGCA------CUAGC---CAAAUGGCCUUCAAUAGCA-CCAAGCAAAUGGCCGCCUGACAC ((((((((........(((.(((((.((((((....))----))))))))).)))..((((..------...((---(....)))......)))).-.........))))))))...... ( -33.70, z-score = -3.08, R) >droGri2.scaffold_14906 6812341 108 - 14172833 GGCGGCCAAAACUAAACGCUUUGAACUUGUAAUCAAUAAAUGGCGAUUGCAUUGC-AGCUGCACGAAGAUCACA---UCUAACAGCUCCAAACUCCACUCUCCACUCCCCCC-------- ((((............)))).......(((((((..........)))))))(((.-(((((...((........---))...))))).))).....................-------- ( -15.80, z-score = 0.36, R) >droMoj3.scaffold_6540 24897470 106 - 34148556 GGCGGCCAAAACUAAACGCUUUGAACUUGUAAUCAAUAAAUGGCGAUUACAUUGCCAGCUGCAUCAAGAUCAAU---UCUAAUA--UCGAAACACCGC-CUCCACUCGCCCC-------- (((((...............((((.((((....((.....(((((((...)))))))..))...)))).))))(---((.....--..)))...))))-)............-------- ( -21.50, z-score = -1.64, R) >droVir3.scaffold_13047 10101312 89 + 19223366 GGCGGCCAAAACUAAACGCUUUGAACUUGUAAUCAAUAAAUGGCGAUUGCAUUGC-AGCUGCACCAAGAUCAAA---UCUAA-----CCAGCUACCCC---------------------- ((..((...........((.(((....(((((((..........)))))))...)-))..))....((((...)---)))..-----...))..))..---------------------- ( -14.80, z-score = 0.45, R) >droWil1.scaffold_181130 1412568 98 - 16660200 GUUGGCCAAAACUAAACGAUUUGAACUUGUAAUCAAUU----GCACCUGCAUUGCCAAUGGCC--ACGCCCAUUUCGCCCACCACUUGCAGCAGCA--ACAGCAAC-------------- ((((.............((((.........))))..((----((..(((((..((.(((((..--....)))))..))........)))))..)))--)))))...-------------- ( -17.50, z-score = 0.85, R) >droPer1.super_7 1039884 99 + 4445127 GGCGGCCAAAACUAAACGUUUUGAACUUGUAAUCGAUU----GCAAUUCAAUUCGCAGCUGCA------UCAGG---UGCCUGGCCUCCAUCUGCACCCCAUCAUUUGAUUC-------- ((.(((((.........(..(((((.((((((....))----)))))))))..)(((.(((..------.))).---))).))))).)).......................-------- ( -26.70, z-score = -1.80, R) >dp4.chr2 27492431 99 - 30794189 GGCGGCCAAAACUAAACGUUUUGAACUUGUAAUCGAUU----GCAAUUCAAUUCGCAGCUGCA------UCAGG---UGCCUGGCCUCCAUCUGCACCCCAUCAUUUGAUUC-------- ((.(((((.........(..(((((.((((((....))----)))))))))..)(((.(((..------.))).---))).))))).)).......................-------- ( -26.70, z-score = -1.80, R) >droAna3.scaffold_13340 21792166 100 + 23697760 GGCGGCCAAAUCUAAACGAUUUGAACUUGUAAUCGAUU----GCAAUUCAAUUUGCAGCUGCA------UUGGCCUUCAGGCCACUUCUCAAGCCAACACAGCCCGCAGC---------- .((((.((((((.....))))))..............(----((((......)))))((((..------(((((.((.(((.....))).)))))))..))))))))...---------- ( -28.20, z-score = -1.63, R) >droEre2.scaffold_4820 769989 106 - 10470090 GGCGGCCAAAUCUAAACGAUUUGAACUUGUAAUCGAUU----GCGAUUCAAUUUGCAGCUGCA------CCAGC---CAAAUGGCCUUCAAUAACC-GCCAACAAAUGGCCGCCUGCAGC ((((((((........(((.(((((.((((((....))----))))))))).)))..((((..------.))))---....((((...........-)))).....))))))))...... ( -32.40, z-score = -2.23, R) >droYak2.chr3R 19224225 113 + 28832112 GGCGGCCAAAUCUAAACGAUUUGAACUUGUAAUCGAUU----GCGAUUCAAUUUGCAGCUGCAUUUGCACCAGCA--CAACUGGCCUUCAAUAACCCGCCAACAAAUGGCCGCCUGCAA- ((((((((........(((.(((((.((((((....))----))))))))).)))..(((((....)))((((..--...)))).............)).......)))))))).....- ( -33.20, z-score = -2.25, R) >droSec1.super_0 18730120 92 + 21120651 GGCGGCCAAAUCUAAACGAUUUGAACUUGUAAUCGAUU----GCGAUUCAAUUUGCAGCUGCA------CCAGC---CAAAUGGCCUUCAAUAGCCGCCUGCCAC--------------- ((((((((((((.....))))))...(((.(((((...----.))))))))..(((....)))------...((---(....)))........))))))......--------------- ( -27.30, z-score = -2.23, R) >droSim1.chr3R 18186666 106 + 27517382 GGCGGCCAAAUCUAAACGAUUUGAACUUGUAAUCGAUU----GCGAUUCAAUUUGCAGCUGCA------CCAGC---CAAAUGGCCUUCAAUAGCU-GCCAGCAAAUGGCCGCCUGCCAC ((((((((.........((.(((((.((((((....))----))))))))).))(((((((..------...((---(....)))......)))))-)).......))))))))...... ( -38.40, z-score = -3.45, R) >consensus GGCGGCCAAAACUAAACGAUUUGAACUUGUAAUCGAUU____GCGAUUCAAUUUGCAGCUGCA______CCAGC___CAAAUGGCCUCCAAUAGCC_CCCAGCAAAUGGCC_________ .(((((((((((.....))))))...(((....))).....................))))).......................................................... ( -9.46 = -9.50 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:31 2011