
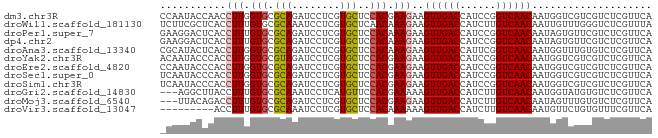
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,359,394 – 18,359,476 |
| Length | 82 |
| Max. P | 0.840740 |

| Location | 18,359,394 – 18,359,476 |
|---|---|
| Length | 82 |
| Sequences | 12 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 84.92 |
| Shannon entropy | 0.34283 |
| G+C content | 0.49439 |
| Mean single sequence MFE | -22.49 |
| Consensus MFE | -13.65 |
| Energy contribution | -13.75 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
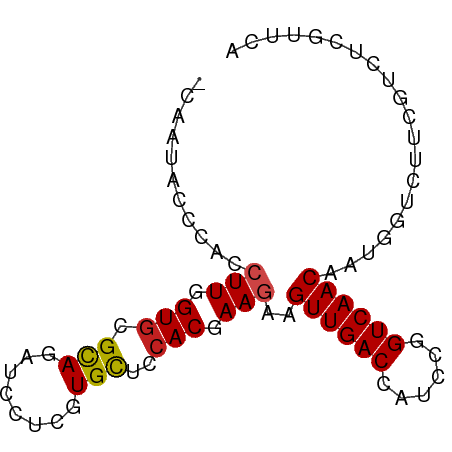
>dm3.chr3R 18359394 82 + 27905053 CCAAUACCAACCUUGGUGCGCAGAUCCUCGUGCUCCACGAAGAAGUUGACCAUCCGGUCAACAAUGGUCGUCGUCUCGUUCA .....((((..(.(((.((((........)))).))).).....(((((((....)))))))..)))).............. ( -23.50, z-score = -1.70, R) >droWil1.scaffold_181130 1393153 82 - 16660200 UCUUCGCUCACCUUUGUGCGCAAAUCCUCGUGCUCAACAAAGAAGUUGACCAUCUUGUCAACAAUUGUUUGGGUCUCGUUUA .....(((((.((((((((((........))))...))))))..((((((......)))))).......)))))........ ( -19.90, z-score = -2.26, R) >droPer1.super_7 1022559 82 + 4445127 GAAGGACUCACCUUUGUGCGCAGAUCCUCGUGCUCCACAAAGAAGUUGACCAUCCGGUCAACAAUAGUGUUCGUCUCGUUCA (((((((.((((((((((.((((.....).)))..)))))))..(((((((....)))))))....)))...))))..))). ( -27.60, z-score = -3.36, R) >dp4.chr2 27475199 82 - 30794189 GAAGGACUCACCUUUGUGCGCAGAUCCUCGUGCUCCACAAAGAAGUUGACCAUCCGGUCAACAAUAGUGUUCGUCUCGUUCA (((((((.((((((((((.((((.....).)))..)))))))..(((((((....)))))))....)))...))))..))). ( -27.60, z-score = -3.36, R) >droAna3.scaffold_13340 21772737 82 + 23697760 CGCAUACUCACCUUGGUGCGCAGAUCCUCGUGCUCCACAAAGAAGUUGACCAUUCGGUCAACAAUGGUUUGUGUCUCGUUCA .(.((((..(((.(((.((((........)))).))).......(((((((....)))))))...)))..)))))....... ( -25.60, z-score = -2.38, R) >droYak2.chr3R 19206389 82 + 28832112 ACAAUACCCACCUUGGUGCGUAGAUCCUCGUGCUCCACGAAGAAGUUGACCAUCCGGUCAACAAUGGUCGUCGUCUCGUUCA ....(((.(((....))).)))((((.(((((...)))))....(((((((....)))))))...))))............. ( -23.30, z-score = -1.58, R) >droEre2.scaffold_4820 752566 82 - 10470090 CCAAUACCCACCUUGGUGCGCAGAUCCUCGUGCUCCACGAAGAAGUUGACCAUCCGGUCAACAAUGGUCGUCGUCUCGUUCA .......(((.(.(((.((((........)))).))).).....(((((((....)))))))..)))............... ( -22.10, z-score = -1.15, R) >droSec1.super_0 18712817 82 + 21120651 UCAAUACCCACCUUGGUGCGCAGAUCCUCGUGCUCCACGAAGAAGUUGACCAUCCGGUCAACAAUGGUCGUCGUCUCGUUCA .......(((.(.(((.((((........)))).))).).....(((((((....)))))))..)))............... ( -22.10, z-score = -1.16, R) >droSim1.chr3R 18168387 82 + 27517382 UCAAUACCCACCUUGGUGCGCAGAUCCUCGUGCUCCACGAAGAAGUUGACCAUCCGGUCAACAAUGGUCGUCGUCUCGUUCA .......(((.(.(((.((((........)))).))).).....(((((((....)))))))..)))............... ( -22.10, z-score = -1.16, R) >droGri2.scaffold_14830 2496012 79 - 6267026 ---AGGCUUACCUUUGUGCGCAAAUCCUCAUGUUCCACGAAAAAGUUGACCAUCUUGUCAACAAUGGUAUGUGUCUCGUUCA ---((((.((((((((((.(((........)))..))))))...((((((......))))))...))))...))))...... ( -19.10, z-score = -2.03, R) >droMoj3.scaffold_6540 19783285 79 + 34148556 ---UUACAGACCUUUGUGCGCAGAUCCUCGUGCUCCACGAAGAAGUUGACCAUCUUGUCAACAAUAGUUUGUGUCUCGUUCA ---.((((((((((((((.((((.....).)))..)))))))..((((((......))))))....)))))))......... ( -23.00, z-score = -2.69, R) >droVir3.scaffold_13047 584539 73 - 19223366 ---------ACCUUUGUGCGCAAAUCCUCGUGCUCCACAAAAAAGUUGACCAUCUUGUCAACAAUGUUCUGUGUUUCGUUCA ---------...((((((.(((........)))..))))))...((((((......)))))).................... ( -14.00, z-score = -1.88, R) >consensus _CAAUACCCACCUUGGUGCGCAGAUCCUCGUGCUCCACGAAGAAGUUGACCAUCCGGUCAACAAUGGUCUUCGUCUCGUUCA ...........(((.(((.(((........)))..))).)))..((((((......)))))).................... (-13.65 = -13.75 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:30 2011