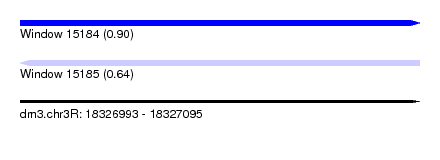
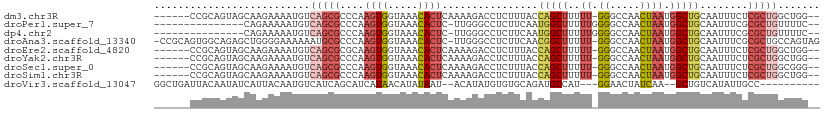
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,326,993 – 18,327,095 |
| Length | 102 |
| Max. P | 0.904673 |

| Location | 18,326,993 – 18,327,095 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.47 |
| Shannon entropy | 0.56853 |
| G+C content | 0.48184 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -13.85 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.904673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
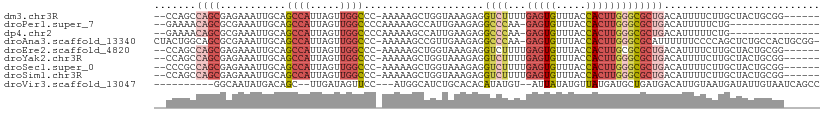
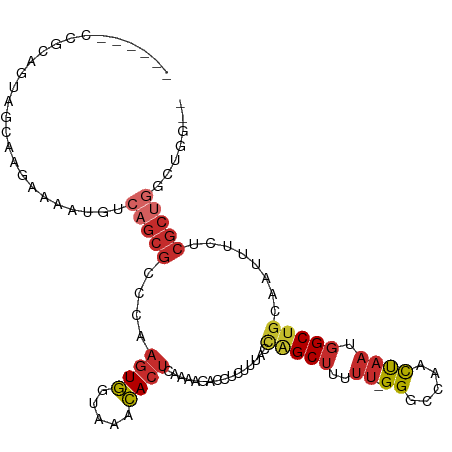
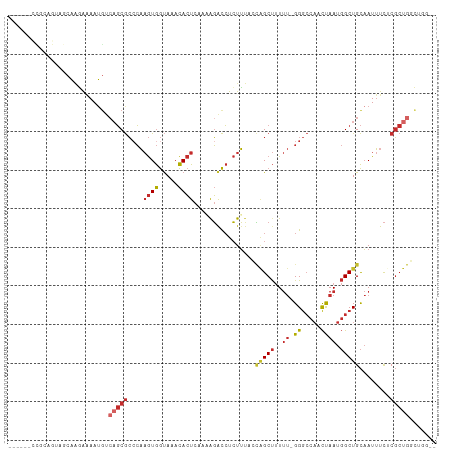
>dm3.chr3R 18326993 102 + 27905053 --CCAGCCAGCGAGAAAUUGCAGCCAUUAGUUGGCCC-AAAAAGCUGGUAAAGAGGUCUUUUGAGUGUUUACCACUUGGGCGCUGACAUUUUCUUGCUACUGCGG------ --((((..(((((((((.((((((((.....))((((-(...((.(((((((..(.((....)).).)))))))))))))))))).)).))))))))).))).).------ ( -38.50, z-score = -2.92, R) >droPer1.super_7 990826 93 + 4445127 --GAAAACAGCGCGAAAUUGCAGCCAUUAGUUGGCCCCAAAAAGCCAUUGAAGAGGCCCAA-GAGUGUUUACCACUUGGGCGCUGACAUUUUUCUG--------------- --(((((((((((....(((..((((.....))))..)))...(((........)))((((-(.(.(....)).)))))))))))....)))))..--------------- ( -28.20, z-score = -2.00, R) >dp4.chr2 27443963 93 - 30794189 --GAAAACAGCGCGAAAUUGCAGCCAUUAGUUGGCCCCAAAAAGCCAUUGAAGAGGCCCAA-GAGUGUUUACCACUUGGGCGCUGACAUUUUUCUG--------------- --(((((((((((....(((..((((.....))))..)))...(((........)))((((-(.(.(....)).)))))))))))....)))))..--------------- ( -28.20, z-score = -2.00, R) >droAna3.scaffold_13340 21737681 108 + 23697760 CUACUGGCAGCGCGAAAUUGCAGCCAUUAGUUGGCCC-AAAAAGCCGUUGAAGAGGCCCAA-GAGUGUUUACCACUUGGGCGCAUUUUUUCCCCAGCUCUGCCACUGCGG- ....((((((.((((((.(((.((((.....))))..-.................((((((-(.(.(....)).))))))))))...))))....)).))))))......- ( -34.80, z-score = -0.58, R) >droEre2.scaffold_4820 718485 102 - 10470090 --CCAGCCAGCGAGAAAUUGCAGCCAUUAGUUGGCCC-AAAAAGCUGGUAAAGAGGUCUUUUGAGUGUUUACCACUUGCGCGCUGACAUUUUCUUGCUACUGCGG------ --((((..(((((((((.((((((((.....))((..-...(((.(((((((..(.((....)).).))))))))))..)))))).)).))))))))).))).).------ ( -31.60, z-score = -1.07, R) >droYak2.chr3R 19171532 102 + 28832112 --CCAGCCAGCGAGAAAUUGCAGCCAUUAGUUGGCCC-AAAAAGCUGGUAAAGAGGUCUUUUGAGUGUUUACCACUUGGGCGCUGACAUUUUCUUGCUACUGCGG------ --((((..(((((((((.((((((((.....))((((-(...((.(((((((..(.((....)).).)))))))))))))))))).)).))))))))).))).).------ ( -38.50, z-score = -2.92, R) >droSec1.super_0 18679830 102 + 21120651 --CCCGCCAGCGAGAAAUUGCAGCCAUUAGUUGGCCC-AAAAAGCUGGUAAAGAGGUCUUUUGAGUGUUUACCACUUGGGCGCUGACAUUUUCUUGCUACUGCGG------ --.((((.(((((((((.((((((((.....))((((-(...((.(((((((..(.((....)).).)))))))))))))))))).)).)))))))))...))))------ ( -42.00, z-score = -4.04, R) >droSim1.chr3R 18134873 102 + 27517382 --CCAGCCAGCGAGAAAUUGCAGCCAUUAGUUGGCCC-AAAAAGCUGGUAAAGAGGUCUUUUGAGUGUUUACCACUUGGGCGCUGACAUUUUCUUGCUACUGCGG------ --((((..(((((((((.((((((((.....))((((-(...((.(((((((..(.((....)).).)))))))))))))))))).)).))))))))).))).).------ ( -38.50, z-score = -2.92, R) >droVir3.scaffold_13047 544678 94 - 19223366 ----------GGCAAUAUGACAGC--UUGAUAGUUCC---AUGGCAUCUGCACACAUAUGU--AUUAUAUGUUAUGAUGCUGAUGACAUUGUAAUGAUAUUGUAAUCAGCC ----------(((..((((..(((--(....)))).)---)))(((((.....(((((((.--..)))))))...)))))((((.(((.(((....))).))).))))))) ( -20.10, z-score = 0.97, R) >consensus __CCAGCCAGCGAGAAAUUGCAGCCAUUAGUUGGCCC_AAAAAGCUGGUAAAGAGGUCUUUUGAGUGUUUACCACUUGGGCGCUGACAUUUUCUUGCUACUGCGG______ .......((((...........((((.....))))....................((((...(((((.....))))))))))))).......................... (-13.85 = -14.77 + 0.92)
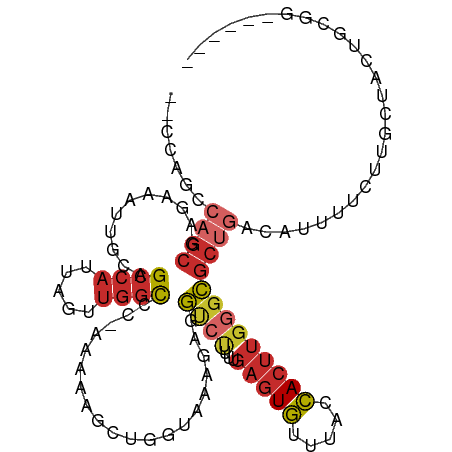
| Location | 18,326,993 – 18,327,095 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.47 |
| Shannon entropy | 0.56853 |
| G+C content | 0.48184 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -11.03 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18326993 102 - 27905053 ------CCGCAGUAGCAAGAAAAUGUCAGCGCCCAAGUGGUAAACACUCAAAAGACCUCUUUACCAGCUUUUU-GGGCCAACUAAUGGCUGCAAUUUCUCGCUGGCUGG-- ------...(((((((.(((((.((.((((((((((((((((((...((....))....)))))))....)))-))))((.....)))))))).))))).))).)))).-- ( -34.20, z-score = -2.51, R) >droPer1.super_7 990826 93 - 4445127 ---------------CAGAAAAAUGUCAGCGCCCAAGUGGUAAACACUC-UUGGGCCUCUUCAAUGGCUUUUUGGGGCCAACUAAUGGCUGCAAUUUCGCGCUGUUUUC-- ---------------..(((((....(((((((((((.(.......).)-))))(((........)))...(((.(((((.....))))).)))....)))))))))))-- ( -31.70, z-score = -2.49, R) >dp4.chr2 27443963 93 + 30794189 ---------------CAGAAAAAUGUCAGCGCCCAAGUGGUAAACACUC-UUGGGCCUCUUCAAUGGCUUUUUGGGGCCAACUAAUGGCUGCAAUUUCGCGCUGUUUUC-- ---------------..(((((....(((((((((((.(.......).)-))))(((........)))...(((.(((((.....))))).)))....)))))))))))-- ( -31.70, z-score = -2.49, R) >droAna3.scaffold_13340 21737681 108 - 23697760 -CCGCAGUGGCAGAGCUGGGGAAAAAAUGCGCCCAAGUGGUAAACACUC-UUGGGCCUCUUCAACGGCUUUUU-GGGCCAACUAAUGGCUGCAAUUUCGCGCUGCCAGUAG -..((((((.((((((((..(((.....(.(((((((.(.......).)-)))))))..)))..)))))))((-((((((.....))))).)))....)))))))...... ( -39.60, z-score = -0.97, R) >droEre2.scaffold_4820 718485 102 + 10470090 ------CCGCAGUAGCAAGAAAAUGUCAGCGCGCAAGUGGUAAACACUCAAAAGACCUCUUUACCAGCUUUUU-GGGCCAACUAAUGGCUGCAAUUUCUCGCUGGCUGG-- ------...(((((((.(((((.(((.(((((....))((((((...((....))....)))))).)))....-.(((((.....)))))))).))))).))).)))).-- ( -30.20, z-score = -1.00, R) >droYak2.chr3R 19171532 102 - 28832112 ------CCGCAGUAGCAAGAAAAUGUCAGCGCCCAAGUGGUAAACACUCAAAAGACCUCUUUACCAGCUUUUU-GGGCCAACUAAUGGCUGCAAUUUCUCGCUGGCUGG-- ------...(((((((.(((((.((.((((((((((((((((((...((....))....)))))))....)))-))))((.....)))))))).))))).))).)))).-- ( -34.20, z-score = -2.51, R) >droSec1.super_0 18679830 102 - 21120651 ------CCGCAGUAGCAAGAAAAUGUCAGCGCCCAAGUGGUAAACACUCAAAAGACCUCUUUACCAGCUUUUU-GGGCCAACUAAUGGCUGCAAUUUCUCGCUGGCGGG-- ------((((..((((.(((((.((.((((((((((((((((((...((....))....)))))))....)))-))))((.....)))))))).))))).)))))))).-- ( -36.90, z-score = -3.15, R) >droSim1.chr3R 18134873 102 - 27517382 ------CCGCAGUAGCAAGAAAAUGUCAGCGCCCAAGUGGUAAACACUCAAAAGACCUCUUUACCAGCUUUUU-GGGCCAACUAAUGGCUGCAAUUUCUCGCUGGCUGG-- ------...(((((((.(((((.((.((((((((((((((((((...((....))....)))))))....)))-))))((.....)))))))).))))).))).)))).-- ( -34.20, z-score = -2.51, R) >droVir3.scaffold_13047 544678 94 + 19223366 GGCUGAUUACAAUAUCAUUACAAUGUCAUCAGCAUCAUAACAUAUAAU--ACAUAUGUGUGCAGAUGCCAU---GGAACUAUCAA--GCUGUCAUAUUGCC---------- (((((((......)))).....(((.((.(.(((((...(((((((..--...)))))))...)))))...---.((....))..--).)).)))...)))---------- ( -18.20, z-score = 0.51, R) >consensus ______CCGCAGUAGCAAGAAAAUGUCAGCGCCCAAGUGGUAAACACUCAAAAGACCUCUUUACCAGCUUUUU_GGGCCAACUAAUGGCUGCAAUUUCUCGCUGGCUGG__ ..........................(((((....((((.....))))...........................(((((.....))))).........)))))....... (-11.03 = -12.28 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:29 2011