| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,326,834 – 18,326,961 |
| Length | 127 |
| Max. P | 0.976144 |

| Location | 18,326,834 – 18,326,961 |
|---|---|
| Length | 127 |
| Sequences | 7 |
| Columns | 136 |
| Reading direction | reverse |
| Mean pairwise identity | 71.55 |
| Shannon entropy | 0.54092 |
| G+C content | 0.51969 |
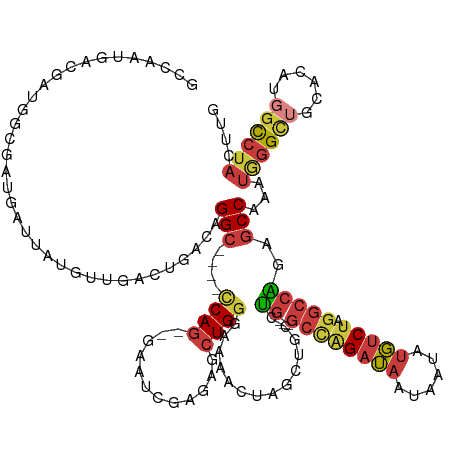
| Mean single sequence MFE | -45.93 |
| Consensus MFE | -16.00 |
| Energy contribution | -14.84 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
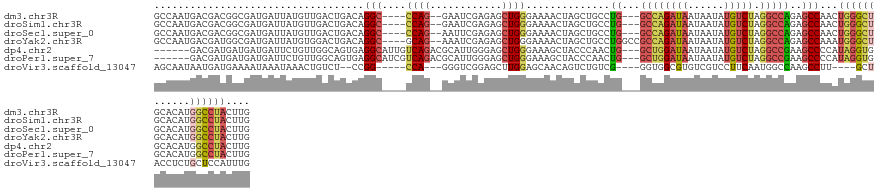
>dm3.chr3R 18326834 127 - 27905053 GCCAAUGACGACGGCGAUGAUUAUGUUGACUGACAGGC----CCAG--GAAUCGAGAGCUGGGAAAACUAGCUGCCUG---GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUG ............(((.(((....((((....))))(((----((((--........((((((.....))))))(((((---((((((((......))))).)))))).))...)))))))...))).)))...... ( -50.80, z-score = -4.36, R) >droSim1.chr3R 18134714 127 - 27517382 GCCAAUGACGACGGCGAUGAUUAUGUUGACUGACAGGC----CCAG--GAAUCGAGAGCUGGGAAAACUAGCUGCCUG---GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUG ............(((.(((....((((....))))(((----((((--........((((((.....))))))(((((---((((((((......))))).)))))).))...)))))))...))).)))...... ( -50.80, z-score = -4.36, R) >droSec1.super_0 18679671 127 - 21120651 GCCAAUGACGACGGCGAUGAUUAUGUUGACUGACAGGC----CCAG--AAUUCGAGAGCUGGGAAAACUAGCUGCCUG---GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAACUGGGCUGCACAUGGCCUACUUG ............(((.(((....((((....))))(((----((((--........((((((.....))))))(((((---((((((((......))))).)))))).))...)))))))...))).)))...... ( -50.60, z-score = -4.64, R) >droYak2.chr3R 19171363 130 - 28832112 GCCAAUGACGAUGGCGAUGAUUAUGUGGACUGACAGGC----GCAG--AAAUCGAGAGCUGGGAAAACUAGCUGCCUGGCCGCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAAAUGGGCUGCACAUGGCCUACUUG ((((.......))))(((.(((((.(((.....(((((----...(--....)...((((((.....)))))))))))....))).))))).....)))(((((((.((((.....)))).....))))))).... ( -45.80, z-score = -2.38, R) >dp4.chr2 27443808 127 + 30794189 ------GACGAUGAUGAUGAUUCUGUUGGCAGUGAGGCAUUGUCAGACGCAUUGGGAGCUGGGAAAGCUACCCAACUG---GCUGGAUAAUAAUAUGUCUAGGCCGAAGCCCCAUAGGUGGCACAUGGCCUACUUG ------((((.(((..(((...(((....))).....)))..)))...((.(((((((((.....)))).)))))...---))............))))(((((((..(((((...)).)))...))))))).... ( -43.20, z-score = -1.48, R) >droPer1.super_7 990671 127 - 4445127 ------GACGAUGAUGAUGAUUCUGUUGGCAGUGAGGCAUCGUCAGACGCAUUGGGAGCUGGGAAAGCUACCCAACUG---GCUGGAUAAUAAUAUGUCUAGGCCGAAGCCCCAUAGGUGGCACAUGGCCUACUUG ------((((.((((((((...(((....))).....))))))))...((.(((((((((.....)))).)))))...---))............))))(((((((..(((((...)).)))...))))))).... ( -45.90, z-score = -2.06, R) >droVir3.scaffold_13047 544560 118 + 19223366 AGCAAUAAUGAUGAAAAUAAAUAAACUGUCU--CCGG-----CCA---GGGUCGGAGCUUGGAGCAACAGUCUGUCG----GGUGGCGUGUCGUCCUUCAAUGGCCAAGCCUU----GCUACCUCUGCUCCAUUUG ...........................(.((--((((-----(..---..)))))))).(((((((((.....)).(----(((((((.((...((......))....))..)----))))))).))))))).... ( -34.40, z-score = -0.47, R) >consensus GCCAAUGACGAUGGCGAUGAUUAUGUUGACUGACAGGC____CCAG__GAAUCGAGAGCUGGGAAAACUAGCUGCCUG___GCCAGAUAAUAAUAUGUCUAGGCCAGAGCCAAAUGGGCUGCACAUGGCCUACUUG ............(((...........................((((............)))).........(((.(.....).)))..........)))(((((((.((((.....)))).....))))))).... (-16.00 = -14.84 + -1.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:27 2011