
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,318,372 – 18,318,477 |
| Length | 105 |
| Max. P | 0.822567 |

| Location | 18,318,372 – 18,318,477 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 67.10 |
| Shannon entropy | 0.60772 |
| G+C content | 0.39272 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
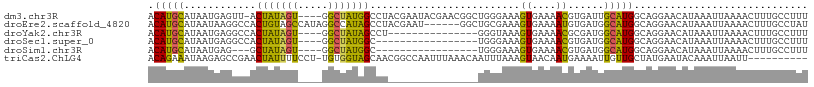
>dm3.chr3R 18318372 105 + 27905053 ACAUGCAUAAUGAGUU-ACUAUAGU----GGCUAUGGCCUACGAAUACGAACGGCUGGGAAAGUGAAAACGUGAUUGCAUGGCAGGAACAUAAAUUAAAACUUUGCCUUU ..(((((..(((((((-((....))----))))..((((..((....))...)))).............)))...)))))((((((...............))))))... ( -21.56, z-score = -0.50, R) >droEre2.scaffold_4820 714984 104 - 10470090 ACAUGCAUAAUAAGGCCACUGUAGCCAUAGGCCAUAGCCUACGAAU------GGCUGCGAAAGUGAAAAUGUGAUGGCAUGGCAGGAACAUAAAUUAAAACUUUGCCUAU ..((((...(((....((((((((((((((((....)))))....)------))))))...))))....)))....))))((((((...............))))))... ( -27.96, z-score = -1.13, R) >droYak2.chr3R 19167989 91 + 28832112 ACAUGCAUAAUGAGGCCACUAUAGU----GGCUAUAGCCU---------------GGGUAAAGUGAAAACGCGAUGGCAUGGCAGGAACAUAAAUUAAAACUUUGCCUUU .(((.....))).((((((....))----)))).......---------------(((((((((......((....))(((.......)))........))))))))).. ( -22.60, z-score = -0.73, R) >droSec1.super_0 18676428 89 + 21120651 ACAUGCAUAAUGAGGCCACUAUAGU----GGCUAUGGC-----------------UGGGAAAGUGAAAACGUGAUGGCAUGGCAGGAACAUAAAUUAAAACUUUGCCUUU ..((((...(((.((((((....))----)))).(.((-----------------(.....))).)...)))....))))((((((...............))))))... ( -21.46, z-score = -0.83, R) >droSim1.chr3R 18131439 86 + 27517382 ACAUGCAUAAUGAG---GCUAUAGU----GGCUAUGGC-----------------UGGGAAAGUGAAAACGUGAUGGCAUGGCAGGAACAUAAAUUAAAACUUUGCCUUU ..((((.......(---(((((((.----..)))))))-----------------)......((....))......))))((((((...............))))))... ( -18.16, z-score = -0.34, R) >triCas2.ChLG4 1883400 99 - 13894384 ACAGAAAUAAGAGCCGAACUAUUUUCCU-UGUGGUAGCAACGGCCAAUUUAAACAAUUUAAAGUAACAAUGAAAAUUGUUGCUAUGAAUACAAAUUAAUU---------- ..........(.((((..(((((.....-...)))))...)))))..........(((((.(((((((((....))))))))).)))))...........---------- ( -21.90, z-score = -3.27, R) >consensus ACAUGCAUAAUGAGGCCACUAUAGU____GGCUAUAGCCU_______________UGGGAAAGUGAAAACGUGAUGGCAUGGCAGGAACAUAAAUUAAAACUUUGCCUUU .(((((............(((((((.....))))))).........................((....))......)))))............................. (-11.15 = -11.23 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:24 2011