| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,294,713 – 18,294,818 |
| Length | 105 |
| Max. P | 0.943232 |

| Location | 18,294,713 – 18,294,818 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Shannon entropy | 0.56148 |
| G+C content | 0.45274 |
| Mean single sequence MFE | -34.41 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.81 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
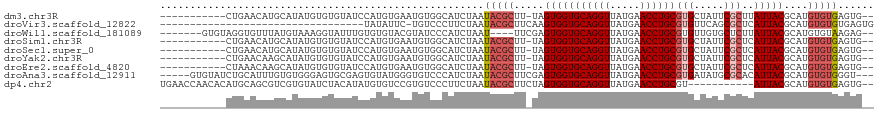
>dm3.chr3R 18294713 105 + 27905053 -----------CUGAACAUGCAUAUGUGUGUAUCCAUGUGAAUGUGGCAUCUAAUACGCUU-UAGUGGUGCAGGUUAUGAACCUGCGUGCUAUUCGCUUAUUACGCAUGUGUGAGUG-- -----------.....(((.((((((((((((.....((((..(((((((......(((..-..)))..((((((.....)))))))))))))))))....)))))))))))).)))-- ( -37.70, z-score = -2.77, R) >droVir3.scaffold_12822 3437640 84 - 4096053 ----------------------------------UAUAUUC-UGUCCCUUCUAAUACGCUUCAAGUGGUGCAGGUUAUGAACCUGCGUGUUCAGCGCUCAUUACGCAUGUGUGUGAGUG ----------------------------------......(-((..((........(((.....)))..((((((.....))))))).)..)))(((((((.(((....)))))))))) ( -21.90, z-score = -0.82, R) >droWil1.scaffold_181089 2768693 106 + 12369635 -------GUGUAGGUGUUUAUGUAAAGGUAUUUGUGUGUACGUAUCCCAUCUAAU----UUCGAGUGGUGCAGGUUAUGAACCUGCGUGUUGUGCUCUUAUUACGCAUGUGUAAGAG-- -------...((((((..((((((.(.(....).)...))))))...))))))..----..........((((((.....))))))........(((((((.((....)))))))))-- ( -23.00, z-score = 0.16, R) >droSim1.chr3R 18103238 105 + 27517382 -----------CUGAACAUGCAUAUGUGUGUAUCCAUGUGAAUGUGGCAUCUAAUACGCUU-UAGUGGUGCAGGUUAUGAACCUGCGUGCUAUUCGCUCAUUACGCAUGUGUGAGUG-- -----------.....(((.((((((((((((.....((((..(((((((......(((..-..)))..((((((.....)))))))))))))))))....)))))))))))).)))-- ( -37.70, z-score = -2.50, R) >droSec1.super_0 18649136 105 + 21120651 -----------CUGAACAUGCAUAUGUGUGUAUCCAUGUGAAUGUGGCAUCUAAUACGCUU-UAGUGGUGCAGGUUAUGAACCUGCGUGCUAUUCGCUCAUUACGCAUGUGUGAGUG-- -----------.....(((.((((((((((((.....((((..(((((((......(((..-..)))..((((((.....)))))))))))))))))....)))))))))))).)))-- ( -37.70, z-score = -2.50, R) >droYak2.chr3R 19140135 105 + 28832112 -----------CUGAACAAGCAUAUGUGUGUAUCCAUGUGAAUGUGGCAUCUAAUACGCUU-UAGUGGUGCAGGUUAUGAACCUGCGUGCUAUUCGCUCAUUACGCAUGUGUGAGUG-- -----------.....((..((((((((((((.....((((..(((((((......(((..-..)))..((((((.....)))))))))))))))))....))))))))))))..))-- ( -37.90, z-score = -2.76, R) >droEre2.scaffold_4820 687176 105 - 10470090 -----------CUAAACAAGCAUAUGUGUGUAUCCAUGUGAAUGUGGCAUCUAAUACGCUU-UAGUGGUGCAGGUUAUGAACCUGCGUGCUAUUCGCUCAUUACGCAUGUGUGAGUG-- -----------.....((..((((((((((((.....((((..(((((((......(((..-..)))..((((((.....)))))))))))))))))....))))))))))))..))-- ( -37.90, z-score = -3.04, R) >droAna3.scaffold_12911 5335513 111 + 5364042 -----GUGUAUCUGCAUUUGUGUGGGAGUGCGAGUGUAUGGGUGUCCCAUCUAAUACGCUUCGAGUGGUGCAGGUUAUGAACCUGCGUGAUAUGCGCACAUUACGCAUGUGUGGGU--- -----....(((..(((.(((((((..(((((((((((((((((...))))).)))))))....((..(((((((.....)))))))..))...))))).))))))).)))..)))--- ( -43.10, z-score = -2.07, R) >dp4.chr2 17008712 106 - 30794189 UGAACCAACACAUGCAGCGUCGUGUAUCUACAUAUGUGUCCGUGUCCCUUCUAAUACGCUUCUAGUGGUGCAGGUUAUGAACCUGCGU-----------AUUACGCAUGUGUGAGUG-- ...((..((((((((.(((.((..(((....)))..))..)))........(((((((((......)))((((((.....))))))))-----------)))).))))))))..)).-- ( -32.80, z-score = -1.53, R) >consensus ___________CUGAACAUGCAUAUGUGUGUAUCCAUGUGAAUGUGGCAUCUAAUACGCUU_UAGUGGUGCAGGUUAUGAACCUGCGUGCUAUUCGCUCAUUACGCAUGUGUGAGUG__ ......................................................(((((.....(((((((((((.....))))))(((.....)))..)))))....)))))...... (-15.10 = -15.81 + 0.71)
| Location | 18,294,713 – 18,294,818 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Shannon entropy | 0.56148 |
| G+C content | 0.45274 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -10.93 |
| Energy contribution | -11.40 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
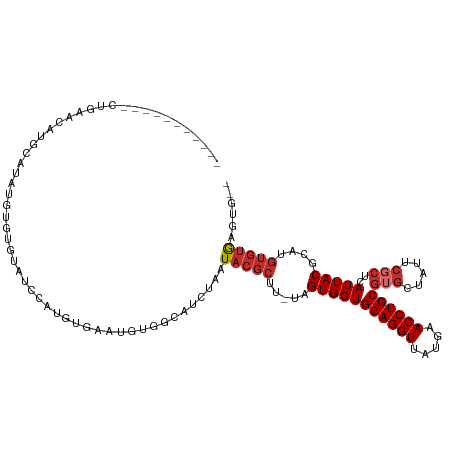
>dm3.chr3R 18294713 105 - 27905053 --CACUCACACAUGCGUAAUAAGCGAAUAGCACGCAGGUUCAUAACCUGCACCACUA-AAGCGUAUUAGAUGCCACAUUCACAUGGAUACACACAUAUGCAUGUUCAG----------- --.......(((((((((....((...(((...((((((.....))))))....)))-..))(((((..(((.........))).))))).....)))))))))....----------- ( -26.30, z-score = -2.55, R) >droVir3.scaffold_12822 3437640 84 + 4096053 CACUCACACACAUGCGUAAUGAGCGCUGAACACGCAGGUUCAUAACCUGCACCACUUGAAGCGUAUUAGAAGGGACA-GAAUAUA---------------------------------- ..(((.....(((.....))).(((((......((((((.....)))))).........))))).......)))...-.......---------------------------------- ( -17.76, z-score = -0.67, R) >droWil1.scaffold_181089 2768693 106 - 12369635 --CUCUUACACAUGCGUAAUAAGAGCACAACACGCAGGUUCAUAACCUGCACCACUCGAA----AUUAGAUGGGAUACGUACACACAAAUACCUUUACAUAAACACCUACAC------- --..........((((((...............((((((.....)))))).(((.((...----....)))))..))))))...............................------- ( -18.70, z-score = -2.46, R) >droSim1.chr3R 18103238 105 - 27517382 --CACUCACACAUGCGUAAUGAGCGAAUAGCACGCAGGUUCAUAACCUGCACCACUA-AAGCGUAUUAGAUGCCACAUUCACAUGGAUACACACAUAUGCAUGUUCAG----------- --.......(((((((((.((.((...(((...((((((.....))))))....)))-..))(((((..(((.........))).)))))...)))))))))))....----------- ( -26.70, z-score = -2.12, R) >droSec1.super_0 18649136 105 - 21120651 --CACUCACACAUGCGUAAUGAGCGAAUAGCACGCAGGUUCAUAACCUGCACCACUA-AAGCGUAUUAGAUGCCACAUUCACAUGGAUACACACAUAUGCAUGUUCAG----------- --.......(((((((((.((.((...(((...((((((.....))))))....)))-..))(((((..(((.........))).)))))...)))))))))))....----------- ( -26.70, z-score = -2.12, R) >droYak2.chr3R 19140135 105 - 28832112 --CACUCACACAUGCGUAAUGAGCGAAUAGCACGCAGGUUCAUAACCUGCACCACUA-AAGCGUAUUAGAUGCCACAUUCACAUGGAUACACACAUAUGCUUGUUCAG----------- --.................(((((.........((((((.....)))))).......-((((((((......(((........)))........))))))))))))).----------- ( -23.64, z-score = -1.09, R) >droEre2.scaffold_4820 687176 105 + 10470090 --CACUCACACAUGCGUAAUGAGCGAAUAGCACGCAGGUUCAUAACCUGCACCACUA-AAGCGUAUUAGAUGCCACAUUCACAUGGAUACACACAUAUGCUUGUUUAG----------- --.......(((.(((((.((.((...(((...((((((.....))))))....)))-..))(((((..(((.........))).)))))...))))))).)))....----------- ( -22.40, z-score = -0.75, R) >droAna3.scaffold_12911 5335513 111 - 5364042 ---ACCCACACAUGCGUAAUGUGCGCAUAUCACGCAGGUUCAUAACCUGCACCACUCGAAGCGUAUUAGAUGGGACACCCAUACACUCGCACUCCCACACAAAUGCAGAUACAC----- ---.....(((((.....))))).((((.....((((((.....))))))..........(((......(((((...))))).....)))............))))........----- ( -28.30, z-score = -2.38, R) >dp4.chr2 17008712 106 + 30794189 --CACUCACACAUGCGUAAU-----------ACGCAGGUUCAUAACCUGCACCACUAGAAGCGUAUUAGAAGGGACACGGACACAUAUGUAGAUACACGACGCUGCAUGUGUUGGUUCA --.(((.((((((((.....-----------..((((((.....)))))).........(((((..........(((.(......).))).........))))))))))))).)))... ( -30.41, z-score = -1.70, R) >consensus __CACUCACACAUGCGUAAUGAGCGAAUAGCACGCAGGUUCAUAACCUGCACCACUA_AAGCGUAUUAGAUGCCACAUUCACAUGGAUACACACAUAUGCAUGUUCAG___________ ...........((((((.....(.......)..((((((.....))))))..........))))))..................................................... (-10.93 = -11.40 + 0.47)

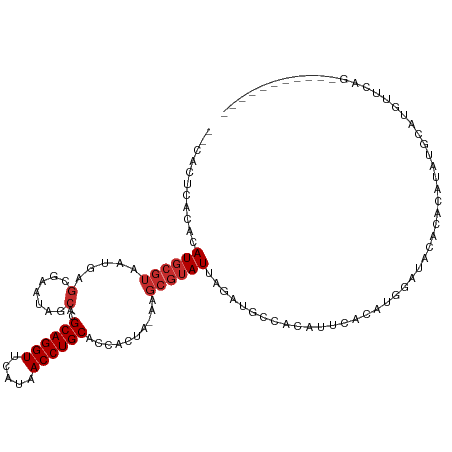
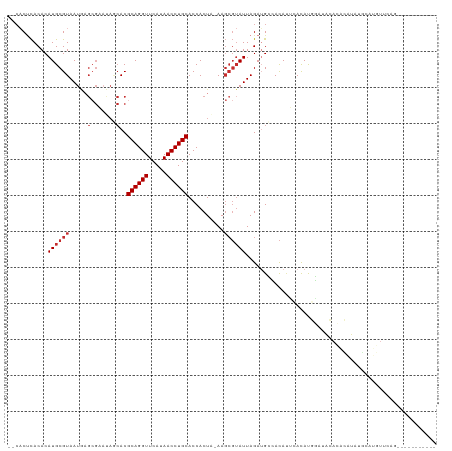
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:19 2011