
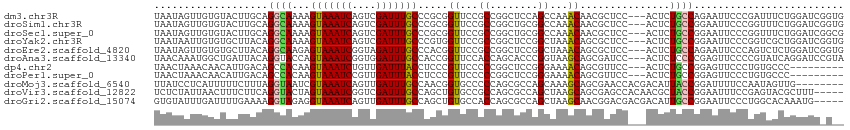
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,272,765 – 18,272,877 |
| Length | 112 |
| Max. P | 0.544205 |

| Location | 18,272,765 – 18,272,877 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.34 |
| Shannon entropy | 0.68297 |
| G+C content | 0.52090 |
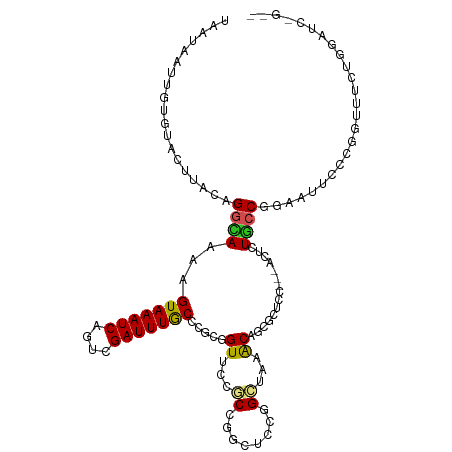
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.21 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18272765 112 - 27905053 UAAUAGUUGUGUACUUGCAGGCAAAAGUAAAUCAGUCGAUUUGCCCGCGGUUCCGCCGGCUCCAGCCAAACAACGCUCC---ACUCUGCCAGAAUUCCCGAUUUCUGGAUCGGUG .....(((((((....)).(((......(((((....)))))(((.(((....))).)))....)))..)))))....(---(((...((((((........))))))...)))) ( -33.80, z-score = -2.04, R) >droSim1.chr3R 18083080 112 - 27517382 UAAUAGUUGUGUACUUGCAGGCAAAAGUAAAUCAGUCGAUUUGCCCGCGGUUCCGCCGGCUGCGGCCAAACAACGCUCC---ACUCUGCCGGAAUUCCCGGUUUCUGGAUCGGUG .....(((((....((((((.(....(((((((....)))))))..(((....))).).))))))....)))))(.(((---(....(((((.....)))))...)))).).... ( -38.30, z-score = -1.55, R) >droSec1.super_0 18628633 112 - 21120651 UAAUAGUUGUGUACUUGCAGGCAAAAGUAAAUCAGUCGAUUUGCCCGCGGUUCCGCCGGCUGCGGCCAAACAACGCUCC---ACUCUGCCGGAAUUCCCGGUUUCUGGAUCGGCG .....(((((....((((((.(....(((((((....)))))))..(((....))).).))))))....)))))(((((---(....(((((.....)))))...))))...)). ( -39.10, z-score = -1.54, R) >droYak2.chr3R 19120750 112 - 28832112 UAAUAAUUGUGUGCUUACAGGCAAAAGUAAAUCAGUCGAUUUGCCCGUGGUUCCGCCGGCUCCGGCUAAACAGCGCUCC---ACUCUGCCGGAAUUCCCGGUCGCUGGAUCGGUG ....(((..((((((....))))...(((((((....))))))).))..))).((((((.((((((....(((.(....---.).)))((((.....))))..)))))))))))) ( -35.90, z-score = -1.00, R) >droEre2.scaffold_4820 667511 112 + 10470090 UAAUAGUUGUGUGCUUACAGGCAAGAGUAAAUCGGUAGAUUUGCCCACGGUUCCGCCGGCUCCGGCUAAACAGCGCUCC---ACUCUGCCAGAAUUCCCAGUCUCUGGAUCGGUG .....(((((..(((....(((..(.(((((((....))))))).).(((.....))))))..)))...)))))....(---(((...(((((..........)))))...)))) ( -31.90, z-score = -0.16, R) >droAna3.scaffold_13340 14829418 112 - 23697760 UAACAAAUGGCUGAUUACAGGUACCAGUAAAUCGGUGGAUUUGCCACCGGUUCCACCAGCACCCGGUAAGCAGCGAUCC---ACUCUCCCCGAGUUCCCCGUAUCAGGAUCCGUA ......(((((((.((((.(((.(..((.(((((((((.....)))))))))..))..).)))..)))).))))(((((---((((.....))))...........)))))))). ( -33.70, z-score = -1.83, R) >dp4.chr2 16988000 103 + 30794189 UAACUAAACAACAUUGACAGCCACAAGUAAAUCUGUUGAUUUACCUCCCGUUCCCCCGGCUCCGGGAAAACAGCGUUCC---ACUCUGCCGGAGUUCCCUGUGCCC--------- ................((((......(((((((....)))))))((((.(((..((((....))))..))).(((....---....))).))))....))))....--------- ( -25.00, z-score = -1.69, R) >droPer1.super_0 6637749 103 - 11822988 UAACUAAACAACAUUGACAGCCACAAGUAAAUCCGUUGAUUUACCUCCCGUUCCCCCGGCUCCGGGAAAACAGCGUUCC---ACUCUGCCGGAGUUCCCUGUGCCC--------- ................((((......(((((((....)))))))((((.(((..((((....))))..))).(((....---....))).))))....))))....--------- ( -23.80, z-score = -1.35, R) >droMoj3.scaffold_6540 812966 107 + 34148556 UUAUCCUCAUUUUUCUUUAGGUAAUCGUAAAUCAGUUGAUUUGCCAACGGUGCCCCCAGCGCCAGCAAAGCAGCGAACCACGACAUUACCGGAUUUUCCAAUAGUUG-------- ...................(((((((((...((.((((.(((((....(((((.....))))).))))).))))))...))))..)))))((.....))........-------- ( -26.30, z-score = -2.85, R) >droVir3.scaffold_12822 3415618 110 + 4096053 UCUCUAUUAACUUUCUUCAGGUACUAGUAAAUCGGUCGAUUUGCCAGCUGUGCCGCCAGCGCCAGCUAAGCAGCGAGCCACAACGCUACCGGAAUUUCCGAGUACGCUUU----- ...................(((((..(((((((....))))))).....)))))((.(((....)))..))(((((((......)))(((((.....))).)).))))..----- ( -28.20, z-score = -0.13, R) >droGri2.scaffold_15074 3107414 110 - 7742996 GUGUAUUUGAUUUUGAAAAGGUAGAGGUAAAUCAGUUGAUUUGCCAGCUGUGCCACCAGCGCCAGCUAAGCAACGGACGACGACAUUGCCGGAAUUCCCUGGCACAAAUG----- ...((((((................((((((((....)))))))).((((......))))(((((..((....(((.(((.....))))))...))..))))).))))))----- ( -31.10, z-score = -1.10, R) >consensus UAAUAAUUGUGUACUUACAGGCAAAAGUAAAUCAGUCGAUUUGCCCGCGGUUCCGCCGGCUCCGGCUAAACAGCGCUCC___ACUCUGCCGGAAUUCCCGGUUUCUGGAUC_G__ ...................((((...(((((((....))))))).....((...((........))...))...............))))......................... (-10.55 = -10.21 + -0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:17 2011