
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,258,259 – 18,258,354 |
| Length | 95 |
| Max. P | 0.500000 |

| Location | 18,258,259 – 18,258,354 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Shannon entropy | 0.42574 |
| G+C content | 0.34943 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -10.57 |
| Energy contribution | -10.41 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
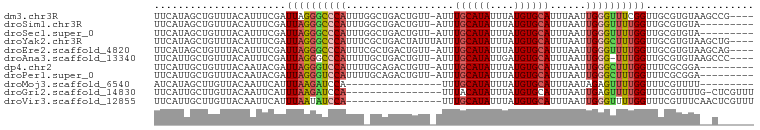
>dm3.chr3R 18258259 95 + 27905053 UUCAUAGCUGUUUACAUUUCGAUUAGGGCCCAUUUGGCUGACUGUU-AUUUGCAUAUUUAUGUGCAUUUAAUUGGGUUUCGGUUGCGUGUAAGCCG---- .........((((((((..((((..(((((((...........(((-(..((((((....))))))..)))))))))))..)))).))))))))..---- ( -27.30, z-score = -2.24, R) >droSim1.chr3R 18068007 90 + 27517382 UUCAUAGCUGUUUACAUUUCGAUUAGGGCCCAUUUGGCUGACUGUU-AUUUGCAUAUUUAUGUGCAUUUAAUUGGGUUUUGGUUGCGUGUA--------- ............(((((..(((((((((((((...........(((-(..((((((....))))))..))))))))))))))))).)))))--------- ( -23.80, z-score = -1.93, R) >droSec1.super_0 18613968 90 + 21120651 UUCAUAGCUGUUUACAUUUCGAUUAGGGCCCAUUUGGCUGACUGUU-AUUUGCAUAUUUAUGUGCAUUUAAUUGGGUUUUGGUUGCGUGUA--------- ............(((((..(((((((((((((...........(((-(..((((((....))))))..))))))))))))))))).)))))--------- ( -23.80, z-score = -1.93, R) >droYak2.chr3R 19105808 96 + 28832112 UUCAUAGCUGUUUACAUUUCGAUUAGGGCCCAUUUCGCUGACUAUUUAUUUGCAUAUUUAUGUGCAUUUAAUUGGGCUUUGGUUGCGUGUAAGCUG---- .........((((((((..(((((((((((((.............(((..((((((....))))))..))).))))))))))))).))))))))..---- ( -30.54, z-score = -3.82, R) >droEre2.scaffold_4820 651470 95 - 10470090 UUCAUAGCUGUUUACAUUUCGAUUAGGGCCCAUUUCGCUGACUGUU-AUUUGCAUAUUUAUGUGCAUUUAAUUGGGUUUUGGUUGCGUGUAAGCAG---- .......((((((((((..(((((((((((((...........(((-(..((((((....))))))..))))))))))))))))).))))))))))---- ( -32.50, z-score = -4.52, R) >droAna3.scaffold_13340 14814296 94 + 23697760 UUCAUUGCUGUUUACAUUUCGAUUAGGGCCCAUUUUGCUGACUGUU-AUUUGCAUAUUGAUGUGCAUUUAAUUGGG-UUUGGUUGCGUGUAAGCCC---- .........((((((((..((((((((.((((...........(((-(..((((((....))))))..))))))))-)))))))).))))))))..---- ( -24.60, z-score = -1.35, R) >dp4.chr2 16971734 90 - 30794189 UUCAUUGCUGUUUACAAUACGAUUAGGGUCCAUUUUGCAGACUGUU-AUUUGCAUAUUUAUGUGCAUUUAAUUGGGCUUUGGUUUCGCGGA--------- ....((((((....))....((((((((((((...........(((-(..((((((....))))))..))))))))))))))))..)))).--------- ( -20.00, z-score = -1.01, R) >droPer1.super_0 6621391 90 + 11822988 UUCAUUGCUGUUUACAAUACGAUUAGGGUCCAUUUUGCAGACUGUU-AUUUGCAUAUUUAUGUGCAUUUAAUUGGGCUUUGGUUUCGCGGA--------- ....((((((....))....((((((((((((...........(((-(..((((((....))))))..))))))))))))))))..)))).--------- ( -20.00, z-score = -1.01, R) >droMoj3.scaffold_6540 27390815 75 + 34148556 AUCAUAGCUUGUUACAAUUCAUUUAAGAUCCA----------------UUUGCAUAUUUAUGUGCAUUUAAUAGAGUUUUGGUUUCGUUUU--------- ((((.(((((((((....((......))....----------------..((((((....))))))..))))).)))).))))........--------- ( -9.40, z-score = -0.33, R) >droGri2.scaffold_14830 48742 83 + 6267026 UUCAUUGCUUGUUACAAUUCAUUUAAGAUCCA----------------UUUACAUAUUUAUGUGCAUUUAAUUGAGUUUUGGUUUCGUUUUG-CUCGUUU ......((.((..((((((((.((((.....(----------------(.(((((....))))).)))))).))))))...))..))....)-)...... ( -6.60, z-score = 1.28, R) >droVir3.scaffold_12855 1669588 84 + 10161210 UUCAUUGCUUGUUACAAUUCAUUUAAUAUCCA----------------UUUGCAUAUUUAUGUGCAUUUAAUUGGGUUUUGGUUUCGUUUCAACUCGUUU ......((..(((..(((....((((.(((((----------------..((((((....))))))......))))).))))....)))..)))..)).. ( -11.00, z-score = -0.72, R) >consensus UUCAUAGCUGUUUACAUUUCGAUUAGGGCCCAUUU_GCUGACUGUU_AUUUGCAUAUUUAUGUGCAUUUAAUUGGGUUUUGGUUGCGUGUA__C______ ......................((((((((((..................((((((....))))))......)))))))))).................. (-10.57 = -10.41 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:16 2011