
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,247,367 – 18,247,461 |
| Length | 94 |
| Max. P | 0.649293 |

| Location | 18,247,367 – 18,247,461 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 62.58 |
| Shannon entropy | 0.66713 |
| G+C content | 0.52858 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -9.27 |
| Energy contribution | -8.02 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
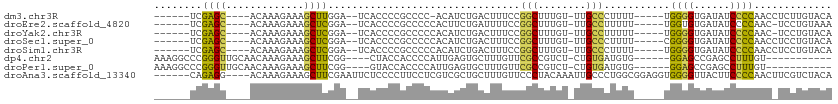
>dm3.chr3R 18247367 94 + 27905053 ------UCGAGC----ACAAAGAAAGCUUGGA--UCACCCCGCCCC-ACAUCUGACUUUCCGGCUUUGU-UUGCCCUUUU-----UGGGGUGAUAUCCCCAACCUCUUGUACA ------..(((.----.((((.((((((.(((--............-...........))))))))).)-)))......(-----(((((......)))))).)))....... ( -22.90, z-score = -0.93, R) >droEre2.scaffold_4820 640094 94 - 10470090 ------UCGAGC----ACAAAGAAAGCUCGGA--UCACCCCGCCCCCACUUCUGAUUUUCCGGCUUUGU-UUGCCUUUUU-----UGGUGUGAUAUCCCCAAC-UCCUGUAAA ------((((((----.........))))))(--((((.((.....((....)).......(((.....-..))).....-----.)).))))).........-......... ( -17.90, z-score = -0.44, R) >droYak2.chr3R 19094782 94 + 28832112 ------UCGAGC----ACAAAGAAAGCUCGGA--UCACCCCGCCCCCACAUCUGACUUUCCGGCUUUGU-UUGCCUUUUU-----UGGGGUGAUAUCCCCAAC-UCCUGUACA ------((((((----.........))))))(--(((((((.....((....)).......(((.....-..))).....-----.)))))))).........-......... ( -25.00, z-score = -1.64, R) >droSec1.super_0 18603157 95 + 21120651 ------UCGAGC----ACAAAGAAAGCUCGGA--UCACCCCGCCCCCACAUCUGACUUUCCGGCUUUGU-UUGCCCUUUU-----CGGGGUGAUAUCCCCAACCUCCUGUACA ------((((((----.........))))))(--((((((((....((....)).......(((.....-..))).....-----)))))))))................... ( -25.80, z-score = -1.91, R) >droSim1.chr3R 18057153 95 + 27517382 ------UCGAGC----ACAAAGAAAGCUCGGA--UCACCCCGCCCCCACAUCUGACUUUCCGGCUUUGU-UUGCCCUUUU-----UGGGGUGAUAUCCCCAACCUCCUGUACA ------..(((.----.((((.(((((.((((--........................))))))))).)-)))......(-----(((((......)))))).)))....... ( -24.66, z-score = -1.51, R) >dp4.chr2 16960645 91 - 30794189 AAAGGCCCGGGUUGCAACAAAGAAAGCUUCGG----CUACCACCCCAUUGAGUGCUUUGUUCGCCGUCU-CUGUGAUGUG------GGAGCCGAGCCUUUGU----------- ((((((.(((.((...........(((....)----)).((((..(((.(((.((.......))...))-).)))..)))------))).))).))))))..----------- ( -31.80, z-score = -1.39, R) >droPer1.super_0 6610274 91 + 11822988 AAAGGCCCGGGUUGCAACAAAGAAAGCUUCGG----GUACCACCCCAUUGAGUGCUUUGUUCGCCGUCU-CUGUGAUGUG------GGAGCCGAGCCUUUGU----------- ((((((.(((.(((((((((((...((((.((----(.....)))....)))).))))))).))((((.-....))))..------.)).))).))))))..----------- ( -32.20, z-score = -1.35, R) >droAna3.scaffold_13340 14803841 103 + 23697760 ------CAGAGG----ACAAAGAAAGCUUCGAAUUCUCCCCUUCCUCGUCGCUGCUUUGUUCCCUACAAAUUGCCCUGGCGGAGGUGGGGUUACUUCCCCAACUUCGUCUACA ------.((.((----((((((..(((..(((.............)))..))).)))))))).))............(((((((.(((((......))))).))))))).... ( -33.92, z-score = -2.35, R) >consensus ______UCGAGC____ACAAAGAAAGCUCGGA__UCACCCCGCCCCCACAUCUGACUUUCCGGCUUUGU_UUGCCCUUUU_____UGGGGUGAUAUCCCCAAC_UCCUGUACA ........((((.............))))................................(((........)))...........((((......))))............. ( -9.27 = -8.02 + -1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:15 2011