
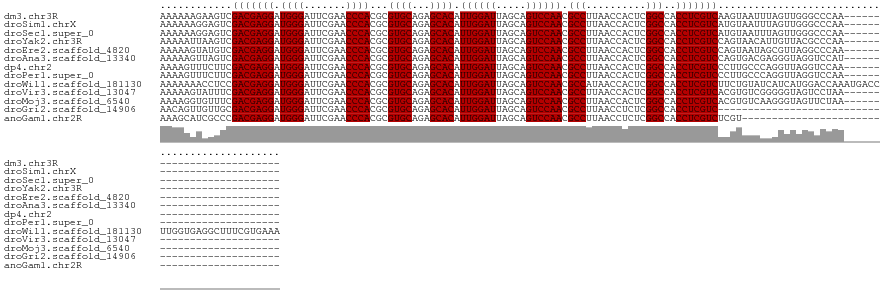
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,222,890 – 18,223,004 |
| Length | 114 |
| Max. P | 0.672435 |

| Location | 18,222,890 – 18,223,004 |
|---|---|
| Length | 114 |
| Sequences | 13 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Shannon entropy | 0.36786 |
| G+C content | 0.53196 |
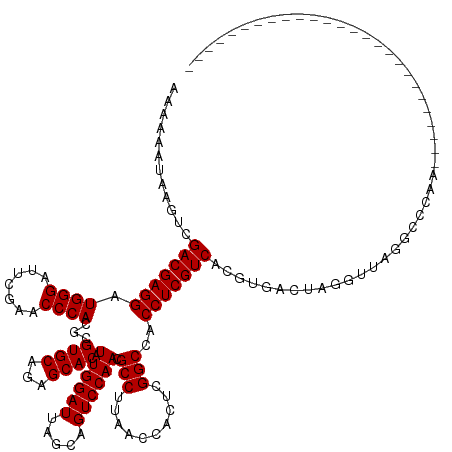
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -28.75 |
| Energy contribution | -28.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18222890 114 + 27905053 AAAAAAGAAGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCAAGUAAUUUAGUUGGGCCCAA-------------------------- ............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................))))))).....................-------------------------- ( -29.30, z-score = -0.18, R) >droSim1.chrX 14667247 114 + 17042790 AAAAAAGGAGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCAUGUAAUUUAGUUGGGCCCAA-------------------------- ......((.((((((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))...((((...)))))))))..-------------------------- ( -32.70, z-score = -0.61, R) >droSec1.super_0 18579104 114 + 21120651 AAAAAAGGAGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCAUGUAAUUUAGUUGGGCCCAA-------------------------- ......((.((((((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))...((((...)))))))))..-------------------------- ( -32.70, z-score = -0.61, R) >droYak2.chr3R 19066627 114 + 28832112 AAAAAUUAAGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCCAGUAACAUUGUUACGCCCAA-------------------------- ............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..(((((...)))))......-------------------------- ( -31.20, z-score = -1.58, R) >droEre2.scaffold_4820 615662 114 - 10470090 AAAAAGUAUGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCCAGUAAUAGCGUUAGGCCCAA-------------------------- .........((((((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..((....))....)))....-------------------------- ( -29.80, z-score = -0.27, R) >droAna3.scaffold_13340 14778494 114 + 23697760 AAAAAGUUAGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCCAGUGACGAGGGUAGGUCCAU-------------------------- .....(((....)))(((..((((.......)))).((((((...)))).((((((.....)))))).))........)))((((.(((((((....)))))))...))))...-------------------------- ( -36.10, z-score = -1.21, R) >dp4.chr2 16931867 114 - 30794189 AAAAGUUUCUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCCCUUGCCCAGGUUAGGUCCAA-------------------------- ............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))(((.((....)).))).....-------------------------- ( -32.80, z-score = -0.68, R) >droPer1.super_0 6581813 114 + 11822988 AAAAGUUUCUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCCCUUGCCCAGGUUAGGUCCAA-------------------------- ............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))(((.((....)).))).....-------------------------- ( -32.80, z-score = -0.68, R) >droWil1.scaffold_181130 12560467 140 + 16660200 AAAAAAACCUCCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCAUAACCACUCGGCCACCUCGUCUUCUGUAUCAUCAUGGACCAAAUGACCUUGGUGAGGCUUUCGUGAAA .......((((.(((((((.((((.......))))...((((...)))).((((((.....)))))).(((..........)))..)))))))................(((((......)))))))))........... ( -38.00, z-score = -0.28, R) >droVir3.scaffold_13047 5200509 114 - 19223366 AAAAAGUAUUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCACGUGUCGGGGGUAGUCCUAA-------------------------- ....................(((((((.(..((((((((((..(((....((((((.....)))))).(((..........)))...)))..))))))).)))..)))))))).-------------------------- ( -34.90, z-score = -0.61, R) >droMoj3.scaffold_6540 26063616 114 + 34148556 AAAAGGUGUUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCACGUGUCAAGGGUAGUUCUAA-------------------------- ....((..(...(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))..)..))..............-------------------------- ( -29.80, z-score = 0.74, R) >droGri2.scaffold_14906 5239067 93 + 14172833 AACAGUUGUUGCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUC----------------------------------------------- ............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))----------------------------------------------- ( -27.60, z-score = -0.66, R) >anoGam1.chr2R 26666066 97 + 62725911 AAAGCAUCGCCCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCUCUCGGCCACCUCGUCUCGU------------------------------------------- ............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))................)))))))....------------------------------------------- ( -28.80, z-score = -0.67, R) >consensus AAAAAAUAAGUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUAACCACUCGGCCACCUCGUCACGUGACUAGGUUAGGCCCAA__________________________ ............(((((((.((((.......))))...((((...)))).((((((.....)))))).(((..........)))..)))))))............................................... (-28.75 = -28.75 + 0.00)

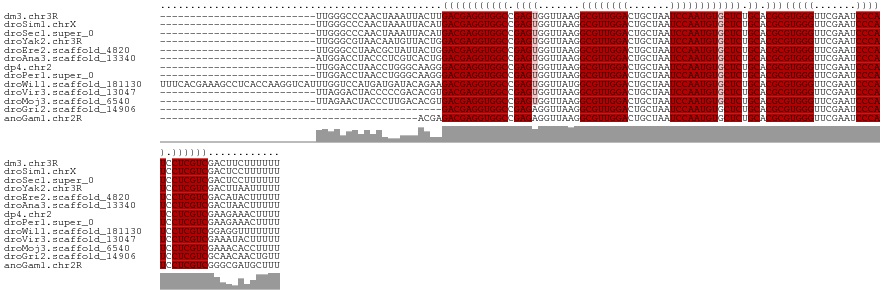
| Location | 18,222,890 – 18,223,004 |
|---|---|
| Length | 114 |
| Sequences | 13 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Shannon entropy | 0.36786 |
| G+C content | 0.53196 |
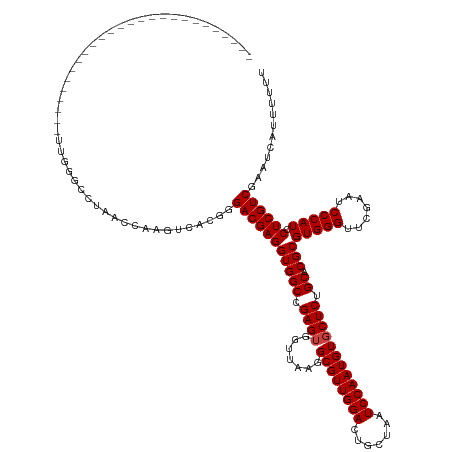
| Mean single sequence MFE | -36.91 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.16 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18222890 114 - 27905053 --------------------------UUGGGCCCAACUAAAUUACUUGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACUUCUUUUUU --------------------------((((......)))).....(((((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).)))))))).......... ( -35.91, z-score = -1.07, R) >droSim1.chrX 14667247 114 - 17042790 --------------------------UUGGGCCCAACUAAAUUACAUGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACUCCUUUUUU --------------------------((((......))))......((((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).)))))))........... ( -35.11, z-score = -0.61, R) >droSec1.super_0 18579104 114 - 21120651 --------------------------UUGGGCCCAACUAAAUUACAUGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACUCCUUUUUU --------------------------((((......))))......((((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).)))))))........... ( -35.11, z-score = -0.61, R) >droYak2.chr3R 19066627 114 - 28832112 --------------------------UUGGGCGUAACAAUGUUACUGGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACUUAAUUUUU --------------------------(((((((((((...)))))..(((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).))))))).)))))..... ( -37.51, z-score = -1.14, R) >droEre2.scaffold_4820 615662 114 + 10470090 --------------------------UUGGGCCUAACGCUAUUACUGGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACAUACUUUUU --------------------------...(((.....))).......(((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).))))))............ ( -34.91, z-score = -0.18, R) >droAna3.scaffold_13340 14778494 114 - 23697760 --------------------------AUGGACCUACCCUCGUCACUGGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGACUAACUUUUU --------------------------.(((((((((.((((((....))))))(((((.((((.......((((((((.......)))))))))))).)).))))))))))))........................... ( -38.71, z-score = -1.18, R) >dp4.chr2 16931867 114 + 30794189 --------------------------UUGGACCUAACCUGGGCAAGGGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAGAAACUUUU --------------------------..........(((.....)))(((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).))))))............ ( -36.51, z-score = 0.04, R) >droPer1.super_0 6581813 114 - 11822988 --------------------------UUGGACCUAACCUGGGCAAGGGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAGAAACUUUU --------------------------..........(((.....)))(((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).))))))............ ( -36.51, z-score = 0.04, R) >droWil1.scaffold_181130 12560467 140 - 16660200 UUUCACGAAAGCCUCACCAAGGUCAUUUGGUCCAUGAUGAUACAGAAGACGAGGUGGCCGAGUGGUUAUGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGGAGGUUUUUUU ......(((((((((((((.((((((((.(((..((......))...))).))))))))...))))..(((((.(((....(.((((((.(((((...)))))..)))))).)....)))....)))))))))))))).. ( -49.10, z-score = -1.66, R) >droVir3.scaffold_13047 5200509 114 + 19223366 --------------------------UUAGGACUACCCCCGACACGUGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAAUACUUUUU --------------------------...((.......))......((((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).)))))))........... ( -34.51, z-score = -0.16, R) >droMoj3.scaffold_6540 26063616 114 - 34148556 --------------------------UUAGAACUACCCUUGACACGUGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAACACCUUUU --------------------------....................((((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).)))))))........... ( -33.71, z-score = -0.30, R) >droGri2.scaffold_14906 5239067 93 - 14172833 -----------------------------------------------GACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGCAACAACUGUU -----------------------------------------------((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))............ ( -35.20, z-score = -1.91, R) >anoGam1.chr2R 26666066 97 - 62725911 -------------------------------------------ACGAGACGAGGUGGCCGAGAGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGGGCGAUGCUUU -------------------------------------------.((.((((((((((((....)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))...))....... ( -37.00, z-score = -1.13, R) >consensus __________________________UUGGGCCUAACCAAGUCACGGGACGAGGUGGCCGAGUGGUUAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAUCAUUUUUU ...............................................(((((((((((.((((.......((((((((.......)))))))))))).)).)))(((((.......))))).))))))............ (-33.00 = -33.16 + 0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:14 2011