| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,222,246 – 18,222,346 |
| Length | 100 |
| Max. P | 0.989470 |

| Location | 18,222,246 – 18,222,346 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.31 |
| Shannon entropy | 0.39444 |
| G+C content | 0.54020 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -33.44 |
| Energy contribution | -32.02 |
| Covariance contribution | -1.42 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
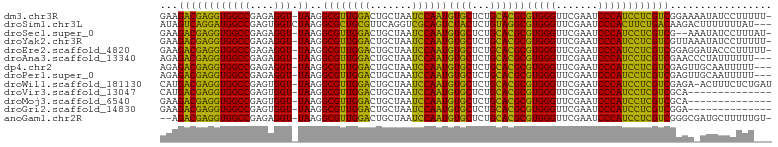
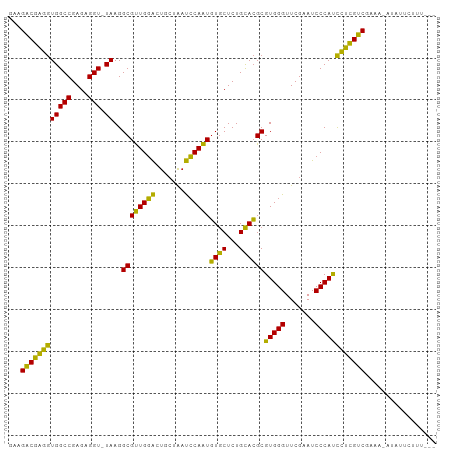
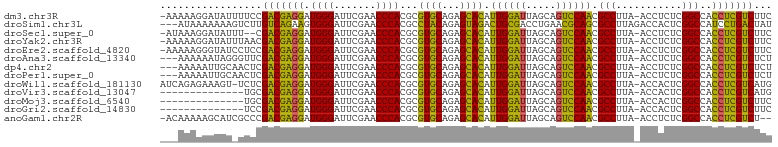
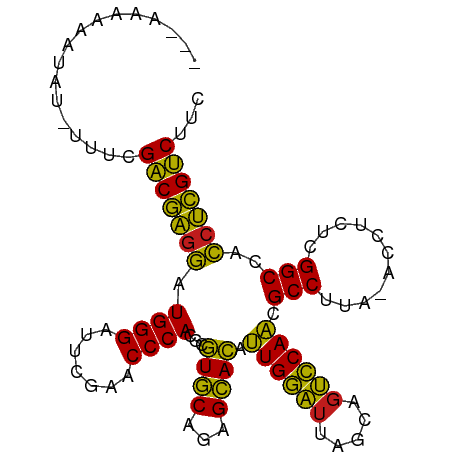
>dm3.chr3R 18222246 100 + 27905053 GAAGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGGAAAAUAUCCUUUUU- ...((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......))))))))))))(((.....))).....- ( -37.30, z-score = -2.44, R) >droSim1.chr3L 15030562 99 - 22553184 AUAGUCAGGAUGGCCGAGUGGUCUAAGGCGCUGCGUUCAGGUCGCAGUCUACUCUGUAGGCGUGGGUUCGAAUCCCACUUCUGACAAGACUUUUUUUAU--- ...(((((((..(((((((((.((..(((.(((....))))))..)).))))))....)))(((((.......))))))))))))..............--- ( -36.10, z-score = -2.54, R) >droSec1.super_0 18578278 98 + 21120651 GAAGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCG--AAAUAUCCUUUAU- ...((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......)))))))))))).--.............- ( -35.40, z-score = -2.29, R) >droYak2.chr3R 19065908 100 + 28832112 GAAGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGUUAAAUAUCCUUUUU- ...((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......))))))))))))................- ( -35.40, z-score = -2.27, R) >droEre2.scaffold_4820 614917 100 - 10470090 GAAGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGGAGGAUACCCUUUUU- ...((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......))))))))))))(((((....)))))..- ( -38.40, z-score = -1.96, R) >droAna3.scaffold_13340 14777878 98 + 23697760 AGAGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAACCCUAUUUUUU--- ...((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......))))))))))))..............--- ( -35.40, z-score = -1.95, R) >dp4.chr2 16931290 98 - 30794189 AGAGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGUUGCAAUUUUU--- ...((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......))))))))))))..............--- ( -35.40, z-score = -1.69, R) >droPer1.super_0 6581235 98 + 11822988 AGAGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGUUGCAAUUUUU--- ...((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......))))))))))))..............--- ( -35.40, z-score = -1.69, R) >droWil1.scaffold_181130 12558288 100 + 16660200 CAUGACGAGGUGGCCGAGUGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAGA-ACUUUCUCUGAU ...(((((((((((.((((...-....((((((((.......)))))))))))).)).)))(((((.......))))).))))))((((-....)))).... ( -36.61, z-score = -1.96, R) >droVir3.scaffold_13047 14022111 87 + 19223366 CAUGACGAGGUGGCCGAGUGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGCA-------------- ...(((((((((((.((((...-....((((((((.......)))))))))))).)).)))(((((.......))))).))))))...-------------- ( -33.51, z-score = -1.64, R) >droMoj3.scaffold_6540 8084199 87 - 34148556 GAAGACGAGGUGGCCGAGUGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGCA-------------- ...(((((((((((.((((...-....((((((((.......)))))))))))).)).)))(((((.......))))).))))))...-------------- ( -33.71, z-score = -1.74, R) >droGri2.scaffold_14830 335854 87 - 6267026 GAAGACGAGGUGGCCGAGUGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGGA-------------- ...(((((((((((.((((...-....((((((((.......)))))))))))).)).)))(((((.......))))).))))))...-------------- ( -33.71, z-score = -1.67, R) >anoGam1.chr2R 36059751 98 - 62725911 --AGACGAGGUGGCCGAGAGGU-UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGGGCGAUGCUUUUUGU- --.((((((((((((....)))-))..((((((((.......))))))((((...))))))(((((.......))))))))))))..((((......))))- ( -36.20, z-score = -1.12, R) >consensus GAAGACGAGGUGGCCGAGAGGU_UAAGGCGUUGGACUGCUAAUCCAAUGUGCUCUGCACGCGUGGGUUCGAAUCCCAUCCUCGUCGAAA_AUAUUCUUU___ ...(((((((..(((....))).....((((((((.......))))))((((...))))))(((((.......))))))))))))................. (-33.44 = -32.02 + -1.42)
| Location | 18,222,246 – 18,222,346 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.31 |
| Shannon entropy | 0.39444 |
| G+C content | 0.54020 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -29.14 |
| Energy contribution | -28.02 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
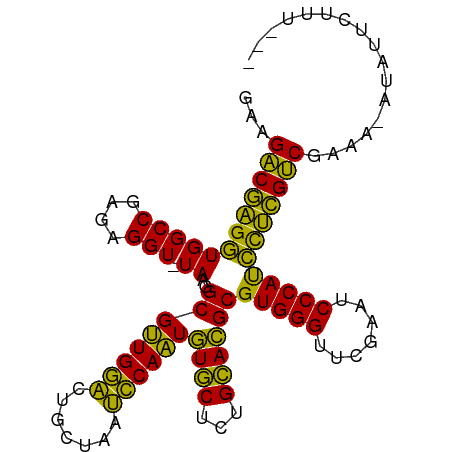
>dm3.chr3R 18222246 100 - 27905053 -AAAAAGGAUAUUUUCCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCUUC -.....(((.....)))(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -31.90, z-score = -2.28, R) >droSim1.chr3L 15030562 99 + 22553184 ---AUAAAAAAAGUCUUGUCAGAAGUGGGAUUCGAACCCACGCCUACAGAGUAGACUGCGACCUGAACGCAGCGCCUUAGACCACUCGGCCAUCCUGACUAU ---..............(((((..(((((.......)))))(((....(((..(.(((((.......))))))..))).((....)))))....)))))... ( -26.90, z-score = -1.96, R) >droSec1.super_0 18578278 98 - 21120651 -AUAAAGGAUAUUU--CGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCUUC -.............--.(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -28.80, z-score = -1.46, R) >droYak2.chr3R 19065908 100 - 28832112 -AAAAAGGAUAUUUAACGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCUUC -................(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -28.80, z-score = -1.63, R) >droEre2.scaffold_4820 614917 100 + 10470090 -AAAAAGGGUAUCCUCCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCUUC -.....(((.....)))(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -30.20, z-score = -1.04, R) >droAna3.scaffold_13340 14777878 98 - 23697760 ---AAAAAAUAGGGUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCUCU ---.......((((..(((.((((.((((.......)))).((((((...)))).((((((.....)))))).))....-.)))))))..).)))....... ( -29.60, z-score = -1.18, R) >dp4.chr2 16931290 98 + 30794189 ---AAAAAUUGCAACUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCUCU ---..............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -28.80, z-score = -1.59, R) >droPer1.super_0 6581235 98 - 11822988 ---AAAAAUUGCAACUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCUCU ---..............(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -28.80, z-score = -1.59, R) >droWil1.scaffold_181130 12558288 100 - 16660200 AUCAGAGAAAGU-UCUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCACUCGGCCACCUCGUCAUG ....((((....-))))(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -32.80, z-score = -2.41, R) >droVir3.scaffold_13047 14022111 87 - 19223366 --------------UGCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCACUCGGCCACCUCGUCAUG --------------...(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -29.30, z-score = -2.09, R) >droMoj3.scaffold_6540 8084199 87 + 34148556 --------------UGCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCACUCGGCCACCUCGUCUUC --------------...(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -28.80, z-score = -1.89, R) >droGri2.scaffold_14830 335854 87 + 6267026 --------------UCCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCACUCGGCCACCUCGUCUUC --------------...(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............)))))))... ( -28.80, z-score = -2.16, R) >anoGam1.chr2R 36059751 98 + 62725911 -ACAAAAAGCAUCGCCCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA-ACCUCUCGGCCACCUCGUCU-- -................(((((((.((((.......)))).((((((...)))).((((((.....)))))).))....-............))))))).-- ( -28.80, z-score = -1.29, R) >consensus ___AAAAAAUAU_UUUCGACGAGGAUGGGAUUCGAACCCACGCGUGCAGAGCACAUUGGAUUAGCAGUCCAACGCCUUA_ACCUCUCGGCCACCUCGUCUUC .................(((((((.((((.......)))).((((((...)))).((((((.....)))))).)).................)))))))... (-29.14 = -28.02 + -1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:12 2011