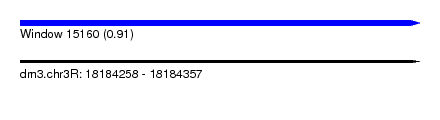
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,184,258 – 18,184,357 |
| Length | 99 |
| Max. P | 0.907289 |

| Location | 18,184,258 – 18,184,357 |
|---|---|
| Length | 99 |
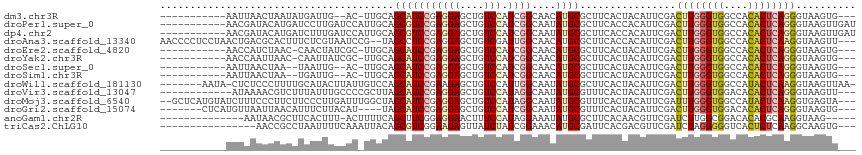
| Sequences | 14 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.09 |
| Shannon entropy | 0.61211 |
| G+C content | 0.48435 |
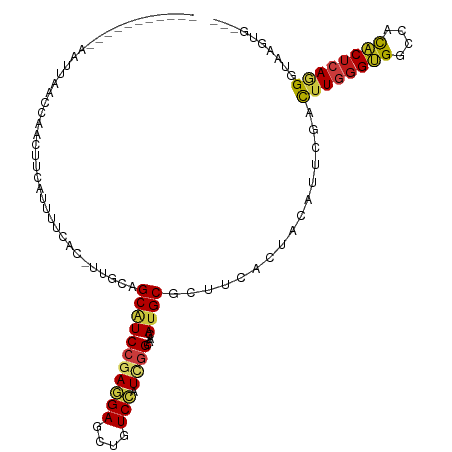
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.18 |
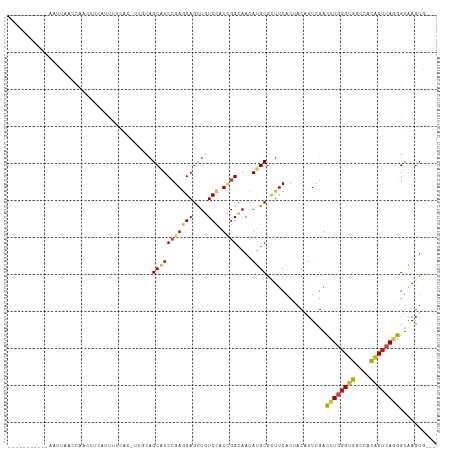
| Combinations/Pair | 1.37 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.907289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18184258 99 + 27905053 -----------AAUUAACUAAUAUGAUUG--AC-UUGCAGCAUCCGAGGAGCUGUCCAUCGGCAACAUGCGCUUCACUACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUG--- -----------..................--((-((((.(((((((((((....))).))))....))))................((((((((....)))))))))))))).--- ( -28.80, z-score = -1.49, R) >droPer1.super_0 5628014 105 - 11822988 -----------AACGAUACAUGAUCCUUGAUCCAUUGCAGCGUCCGAGGAGCUGUCCAUCGGCAAUAUGCGCUUCACCACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUUGAU -----------..((((....((((...))))..((((((((((((((((....))).))))......))))).............((((((((....)))))))))))))))).. ( -28.10, z-score = 0.33, R) >dp4.chr2 14245154 105 + 30794189 -----------AACGAUACAUGAUCUUUGAUCCAUUGCAGCGUCCGAGGAGCUGUCCAUCGGCAAUAUGCGCUUCACCACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUUGAU -----------(((..(((.......((((.....((.((((((((((((....))).))))......))))).))......))))((((((((....))))))))))).)))... ( -28.10, z-score = 0.18, R) >droAna3.scaffold_13340 9165493 111 + 23697760 AACCCCUCCUAACUGACGCACUUUCUCGUAAUCCG--UAGCCUCCGAGGAGCUGUCGAUUGGCAACAUGCGCUUCACCACAUUCGACUUGGGUGGCCACACUCAAGGUAAGUU--- ..........((((.((((.........(((((..--((((.((....))))))..)))))((.....))))..............((((((((....)))))))))).))))--- ( -27.30, z-score = 0.16, R) >droEre2.scaffold_4820 576757 100 - 10470090 -----------AACCAUCUAAC-CAACUAUCGC-UUGCAGCAUCCGAGGAGCUGUCCAUCGGCAACAUGCGCUUCACUACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUG--- -----------...........-.......(((-((((.(((((((((((....))).))))....))))................((((((((....)))))))))))))))--- ( -30.00, z-score = -1.68, R) >droYak2.chr3R 19027878 100 + 28832112 -----------AACCAAUUAAC-CAAUUAUCGC-UUGCAGCAUCCGAGGAGCUGUCCAUCGGCAACAUGCGCUUCACUACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUG--- -----------...........-.......(((-((((.(((((((((((....))).))))....))))................((((((((....)))))))))))))))--- ( -30.00, z-score = -1.87, R) >droSec1.super_0 18540428 97 + 21120651 -----------AAUUAACUAA--UAAUUG--AC-UUGCAGCAUCCGAGGAGCUGUCCAUCGGCAACAUGCGCUUCACUACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUG--- -----------..........--......--((-((((.(((((((((((....))).))))....))))................((((((((....)))))))))))))).--- ( -28.80, z-score = -1.85, R) >droSim1.chr3R 17993286 97 + 27517382 -----------AAUUAACUAA--UGAUUG--AC-UUGCAGCAUCCGAGGAGCUGUCCAUCGGCAACAUGCGCUUCACUACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUG--- -----------..........--......--((-((((.(((((((((((....))).))))....))))................((((((((....)))))))))))))).--- ( -28.80, z-score = -1.42, R) >droWil1.scaffold_181130 12330769 107 + 16660200 -------AAUA-CUCUCCCUUUUGCAUACUUAUUGUCCAGCAUCCGAAGAGCUGUCCAUUGGCAAUAUGCGCUUCACUACAUUCGACUUGGGUGGCCAUACUCAAGGUAAGUUAA- -------..((-(..........(((((.(((.((..((((.((....))))))..)).)))...)))))................((((((((....)))))))))))......- ( -22.30, z-score = 0.04, R) >droVir3.scaffold_13047 10932606 101 - 19223366 ------------AUAAAACGUCUUUAUUUGCCCCGCUUAGCAUCCGAGGAGCUGUCCAUAGGCAAUAUGCGUUUCACUACAUUCGACUUGGGUGGACACACUCAGGGUAAGUU--- ------------...((((((......(((((..(..((((.((....))))))..)...)))))...))))))............((((((((....)))))))).......--- ( -26.60, z-score = -0.93, R) >droMoj3.scaffold_6540 27901075 111 + 34148556 --GCUCAUGUAUCUUUCCCUUCUUCCCUUGAUUUGGCUAGCAUCCGAGGAGCUGUCCAUAGGCAAUAUGCGUUUCACUACAUUCGAUUUGGGUGGCCAUACUCAAGGUGAGUA--- --...................((..((((((..((((...((((((((((....)))....((.....)).................)))))))))))...))))))..))..--- ( -27.50, z-score = 0.16, R) >droGri2.scaffold_15074 1319398 102 + 7742996 -------CUCAUGUUAAUUAACAUUUCUUACAU----UAGCAUCCGAGGAGCUGUCCAUCGGCAAUAUGCGUUUCACUACAUUCGACUUGGGUGGACACACUCAGGGUAAGUG--- -------...(((((....)))))..(((((..----..(((((((((((....))).))))....))))................((((((((....)))))))))))))..--- ( -27.80, z-score = -2.40, R) >anoGam1.chr2R 51551626 96 + 62725911 --------------AAUAACGCUUCACUUU-ACUUUUCAGCUUCGGAGGAACUUUCCAUAGGAAAUAUGCGCUUCACAACGUUCGAUCUUGGCGGACACACGCAAGGUAAG----- --------------..............((-(((((.(((..((((((((....)))....(((........)))......)))))..)))(((......)))))))))).----- ( -16.70, z-score = 0.08, R) >triCas2.ChLG10 2317863 97 + 8806720 ----------------AACCGCCUAAUUUUCAAAUUACAGCGUCGGAAGAGUUAUCUAUCGGAAACAUGCGAUUCACGACGUUCGAUCUAGGGGGUCACUCUCAAGGCAAGUG--- ----------------....((((..............((((((((((((........))(....)......))).))))))).((((.....)))).......)))).....--- ( -23.00, z-score = -0.76, R) >consensus ___________AAUUAACCAACUUCAUUUUCAC_UUGCAGCAUCCGAGGAGCUGUCCAUCGGCAACAUGCGCUUCACUACAUUCGACUUGGGUGGCCACACUCAGGGUAAGUG___ .......................................(((((((((((....))).))))....))))................((((((((....)))))))).......... (-16.75 = -16.94 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:07 2011