| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,170,592 – 18,170,707 |
| Length | 115 |
| Max. P | 0.938185 |

| Location | 18,170,592 – 18,170,707 |
|---|---|
| Length | 115 |
| Sequences | 8 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 75.75 |
| Shannon entropy | 0.46772 |
| G+C content | 0.55385 |
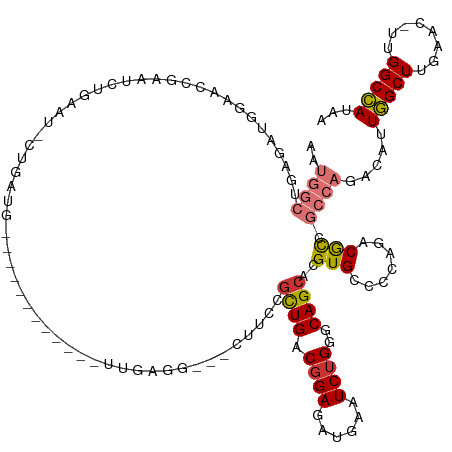
| Mean single sequence MFE | -43.91 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.938185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
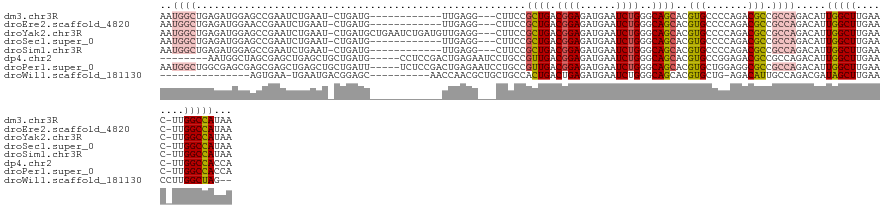
>dm3.chr3R 18170592 115 + 27905053 AAUGGCUGAGAUGGAGCCGAAUCUGAAU-CUGAUG------------UUGAGG---CUUCCGCUGACGGAGAUGAAUCUGGGCAGCACGUGCCCCAGACGCCGCCAGACAUUGGCUUGAAC-UUGGCCAUAA .(((((..((.(.(((((((.((((..(-(.....------------..))((---((((((....))))).....((((((..((....)))))))).)))..))))..))))))).).)-)..))))).. ( -47.40, z-score = -3.79, R) >droEre2.scaffold_4820 563179 115 - 10470090 AAUGGCUGAGAUGGAACCGAAUCUGAAU-CUGAUG------------UUGAGG---CUUCCGCUGACGGAGAUGAAUCUGGGCAGCACGUGCCCCAGACGCCGCCAGACAUUGGCUUGAAC-UUGGCCAUAA .(((((..((.(.((.((((.((((..(-(.....------------..))((---((((((....))))).....((((((..((....)))))))).)))..))))..)))).)).).)-)..))))).. ( -40.80, z-score = -2.37, R) >droYak2.chr3R 19013856 127 + 28832112 AAUGGCUGAGAUGGAGCCGAAUCUGAAU-CUGAUGCUGAAUCUGAUGUUGAGG---CUUCCGCUGACGGAGAUGAAUCUGGGCAGCACGUGCCCCAGACGCCGCCAGACAUUGGCUUGAAC-UUGGCCAUAA .(((((..((.(.(((((((.((((.((-(.(((.....))).))).....((---((((((....))))).....((((((..((....)))))))).)))..))))..))))))).).)-)..))))).. ( -51.20, z-score = -4.02, R) >droSec1.super_0 18525671 115 + 21120651 AAUGGCUGAGAUGGAGCCGAAUCUGAAU-CUGAUG------------UUGAGG---CUUCCGCUGACGGAGAUGAAUCUGGGCAGCACGUGCCCCAGACGCCGCCAGACAUUGGCUUGAAC-UUGGCCAUAA .(((((..((.(.(((((((.((((..(-(.....------------..))((---((((((....))))).....((((((..((....)))))))).)))..))))..))))))).).)-)..))))).. ( -47.40, z-score = -3.79, R) >droSim1.chr3R 17978529 115 + 27517382 AAUGGCUGAGAUGGAGCCGAAUCUGAAU-CUGAUG------------UUGAGG---CUUCCGCUGACGGAGAUGAAUCUGGGCAGCACGUGCCCCAGACGCCGCCAGACAUUGGCUUGAAC-UUGGCCAUAA .(((((..((.(.(((((((.((((..(-(.....------------..))((---((((((....))))).....((((((..((....)))))))).)))..))))..))))))).).)-)..))))).. ( -47.40, z-score = -3.79, R) >dp4.chr2 12324262 118 + 30794189 --------AAUGGCUAGCGAGCUGAGCUGCUGAUG-----CCUCCGACUGAGAAUCCUGCCGUUGACGGAGAUGAAUCUGGGCAGCACGUGCCGGAGACGCCGCCAGACAUUGGCUUGAAC-UUGGCCACCA --------..((((((.(((((..((((((((((.-----.(((((.(...(........)...).)))))....)))..))))))..(((.((....)).)))......)..)))))...-.))))))... ( -41.50, z-score = -1.12, R) >droPer1.super_0 3677167 126 - 11822988 AAUGGCUGGCGAGCGAGCGAGCUGAGCUGCUGAUU-----UCUCCGACUGAGAAUCCUGCCGUUGACGGAGAUGAAUCUGGGCAGCACGUGCUGGAGGCGCCGCCAGACAUUGGCUUGAAC-UUGGCCACCA ((((.((((((.(((..(.(((((.((((((((((-----((((((.(...(........)...).))))))..))))..)))))).)).))).)...))))))))).))))((((.....-..)))).... ( -51.50, z-score = -1.96, R) >droWil1.scaffold_181130 12317823 103 + 16660200 ---------------AGUGAA-UGAAUGACGGAGC----------AACCAACGCUGCUGCCACUGACUGAGAUGAAUCUGGGCAGCACGUGCUG-AGACAUUGCCAGACGAUAGCUUGAACCUUGGCUAG-- ---------------......-..((((.(..(((----------.....(((.(((((((...((..........))..))))))))))))).-.).)))).........(((((........))))).-- ( -24.10, z-score = 0.25, R) >consensus AAUGGCUGAGAUGGAACCGAAUCUGAAU_CUGAUG____________UUGAGG___CUUCCGCUGACGGAGAUGAAUCUGGGCAGCACGUGCCCCAGACGCCGCCAGACAUUGGCUUGAAC_UUGGCCAUAA ..((((.......................................................((((.((((......))))..))))..(((.......))).)))).....(((((........)))))... (-18.05 = -18.68 + 0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:05 2011