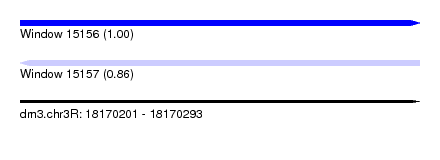
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,170,201 – 18,170,293 |
| Length | 92 |
| Max. P | 0.999058 |

| Location | 18,170,201 – 18,170,293 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.80 |
| Shannon entropy | 0.55782 |
| G+C content | 0.44703 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18170201 92 + 27905053 -------------CAAAAUCCCAGACUCCAGUGCCCAUAACC-CC--UGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA -------------.......((((.(.((((.(........)-.)--))))))))..........(((((((.(((((......))))).)))))))........... ( -23.30, z-score = -2.77, R) >droAna3.scaffold_13340 9152684 86 + 23697760 --------------------CAGAGUCCUAGCCCACCUCCACCCU--CCGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA --------------------..((....(((((..((........--..))..)))))....)).(((((((.(((((......))))).)))))))........... ( -22.70, z-score = -3.40, R) >droEre2.scaffold_4820 562807 91 - 10470090 --------------CAAAUCCCAGACUCCAGUGCCCAUAACC-CC--UGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA --------------......((((.(.((((.(........)-.)--))))))))..........(((((((.(((((......))))).)))))))........... ( -23.30, z-score = -2.74, R) >droYak2.chr3R 19013452 105 + 28832112 CACCCCCUCCCCCAAAAAUCCCAGACUCUAGUGCCCAUAACC-CU--UGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA ............................(((((((((.....-..--)))))..)))).......(((((((.(((((......))))).)))))))........... ( -24.20, z-score = -3.28, R) >droSec1.super_0 18525291 92 + 21120651 -------------CCAAAUCCCAGACUCCAGUGCCCAUAACC-CC--UGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA -------------.......((((.(.((((.(........)-.)--))))))))..........(((((((.(((((......))))).)))))))........... ( -23.30, z-score = -2.71, R) >droSim1.chr3R 17978150 92 + 27517382 -------------CCAAAUCCCAGACUCCAGUGCCCAUAACC-CC--UGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA -------------.......((((.(.((((.(........)-.)--))))))))..........(((((((.(((((......))))).)))))))........... ( -23.30, z-score = -2.71, R) >droGri2.scaffold_15074 1304666 92 + 7742996 --------------CCCAGCGCGACCACCCGCCUGCGUCUCUGCC--CGGAAAGCCUGAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA --------------..(((.(((((((......)).)))(((...--.)))..))))).......(((((((.(((((......))))).)))))))........... ( -22.70, z-score = -1.80, R) >droMoj3.scaffold_6540 27886856 79 + 34148556 -------------------------UAAAGUGUUGCGUCU--GCC--CUCAGCGGCUGAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA -------------------------.......(((((((.--(((--......))).))......(((((((.(((((......))))).)))))))....))))).. ( -20.70, z-score = -1.74, R) >droVir3.scaffold_13047 10917402 79 - 19223366 -------------------------AAAAGUGUUGCGUCU--GCC--CUCAGCGGCUGAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA -------------------------.......(((((((.--(((--......))).))......(((((((.(((((......))))).)))))))....))))).. ( -20.70, z-score = -1.69, R) >droWil1.scaffold_181130 12317434 100 + 16660200 --------UAGGGAGAAAGGGCUGGUAGUAGUAUGAGUAAGAGUUGGCAGCCAGGGCUAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA --------..((.((....(((((.(((...............))).)))))....))....)).(((((((.(((((......))))).)))))))........... ( -23.76, z-score = -1.64, R) >droPer1.super_0 3676550 89 - 11822988 -----------------GUCCCAUCCCUGCUCCACCAUACCGACC--AGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA -----------------...(((.(((((.((.........)).)--)))).)))..........(((((((.(((((......))))).)))))))........... ( -25.50, z-score = -3.55, R) >dp4.chr2 12323678 89 + 30794189 -----------------UUCCCAUCCCUGCUCCACCAUACCGACC--AGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA -----------------...(((.(((((.((.........)).)--)))).)))..........(((((((.(((((......))))).)))))))........... ( -25.50, z-score = -3.76, R) >consensus _______________AAAUCCCAGACUCCAGUGCCCAUAACC_CC__CGGGCUGGCUAAAAAUCAUAUGCAUCACGUGAUUAAACGCGUCAUGCAUGAAAAUGUAAAA .................................................................(((((((.(((((......))))).)))))))........... (-13.90 = -13.90 + 0.00)

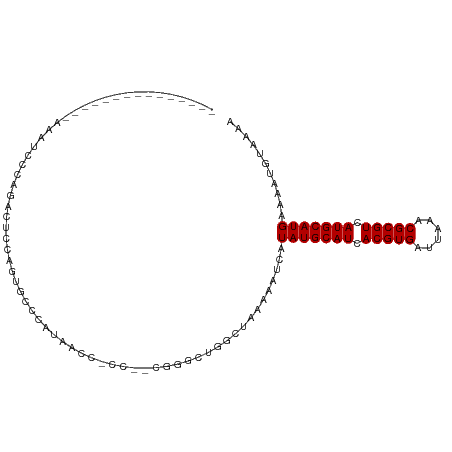

| Location | 18,170,201 – 18,170,293 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.80 |
| Shannon entropy | 0.55782 |
| G+C content | 0.44703 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18170201 92 - 27905053 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCA--GG-GGUUAUGGGCACUGGAGUCUGGGAUUUUG------------- ........(..(((((((.(((.(......).))).))))))..((((((..(((.(((..--..-)))....)))...)))))).)..).....------------- ( -24.70, z-score = -1.25, R) >droAna3.scaffold_13340 9152684 86 - 23697760 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCGG--AGGGUGGAGGUGGGCUAGGACUCUG-------------------- ............((((((.(((.(......).))).))))))..((.((((((((.(((..--........))).)))))))).))..-------------------- ( -26.60, z-score = -2.22, R) >droEre2.scaffold_4820 562807 91 + 10470090 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCA--GG-GGUUAUGGGCACUGGAGUCUGGGAUUUG-------------- .....((..(((((((((.(((.(......).))).))))))..((((((..(((.(((..--..-)))....)))...))))))..)))..))-------------- ( -25.70, z-score = -1.57, R) >droYak2.chr3R 19013452 105 - 28832112 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCA--AG-GGUUAUGGGCACUAGAGUCUGGGAUUUUUGGGGGAGGGGGUG .....((((..(((((((.(((.(......).))).))))))...........((..((((--((-((((.(((((......))))).)))))))))))).)..)))) ( -33.00, z-score = -2.65, R) >droSec1.super_0 18525291 92 - 21120651 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCA--GG-GGUUAUGGGCACUGGAGUCUGGGAUUUGG------------- .....((..(((((((((.(((.(......).))).))))))..((((((..(((.(((..--..-)))....)))...))))))..)))..)).------------- ( -27.00, z-score = -1.80, R) >droSim1.chr3R 17978150 92 - 27517382 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCA--GG-GGUUAUGGGCACUGGAGUCUGGGAUUUGG------------- .....((..(((((((((.(((.(......).))).))))))..((((((..(((.(((..--..-)))....)))...))))))..)))..)).------------- ( -27.00, z-score = -1.80, R) >droGri2.scaffold_15074 1304666 92 - 7742996 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUCAGGCUUUCCG--GGCAGAGACGCAGGCGGGUGGUCGCGCUGGG-------------- ........((((((((((.(((.(......).))).))))))...........(((.....--))).))))(.(((((((....)))).))).)-------------- ( -23.70, z-score = 0.21, R) >droMoj3.scaffold_6540 27886856 79 - 34148556 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUCAGCCGCUGAG--GGC--AGACGCAACACUUUA------------------------- ............((((((.(((.(......).))).))))))..........(((......--)))--...............------------------------- ( -16.60, z-score = -0.57, R) >droVir3.scaffold_13047 10917402 79 + 19223366 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUCAGCCGCUGAG--GGC--AGACGCAACACUUUU------------------------- ............((((((.(((.(......).))).))))))..........(((......--)))--...............------------------------- ( -16.60, z-score = -0.59, R) >droWil1.scaffold_181130 12317434 100 - 16660200 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUAGCCCUGGCUGCCAACUCUUACUCAUACUACUACCAGCCCUUUCUCCCUA-------- ............((((((.(((.(......).))).))))))..............(((((.......................)))))...........-------- ( -16.80, z-score = -2.28, R) >droPer1.super_0 3676550 89 + 11822988 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCU--GGUCGGUAUGGUGGAGCAGGGAUGGGAC----------------- ............((((((.(((.(......).))).))))))...........(((.((((--(.((.(.....).)).))))).)))...----------------- ( -27.60, z-score = -2.03, R) >dp4.chr2 12323678 89 - 30794189 UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCU--GGUCGGUAUGGUGGAGCAGGGAUGGGAA----------------- .........(((((((((.(((.(......).))).))))))...........(((.((((--(.((.(.....).)).))))).))))))----------------- ( -27.70, z-score = -1.87, R) >consensus UUUUACAUUUUCAUGCAUGACGCGUUUAAUCACGUGAUGCAUAUGAUUUUUAGCCAGCCCA__GG_GGUUAUGGGCACUGGAGUCUGGGAUUU_______________ ............((((((.(((.(......).))).)))))).................................................................. (-11.80 = -11.80 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:04 2011