| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,290,681 – 8,290,806 |
| Length | 125 |
| Max. P | 0.996861 |

| Location | 8,290,681 – 8,290,806 |
|---|---|
| Length | 125 |
| Sequences | 13 |
| Columns | 146 |
| Reading direction | forward |
| Mean pairwise identity | 70.93 |
| Shannon entropy | 0.60723 |
| G+C content | 0.53471 |
| Mean single sequence MFE | -44.37 |
| Consensus MFE | -29.82 |
| Energy contribution | -29.71 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
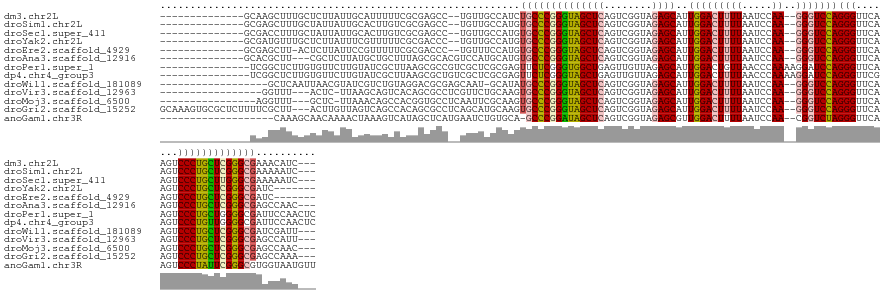
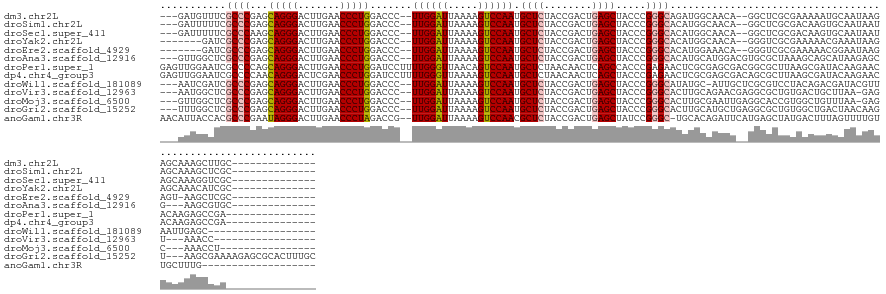
>dm3.chr2L 8290681 125 + 23011544 --------------GCAAGCUUUGCUCUUAUUGCAUUUUUCGCGAGCC--UGUUGCCAUCUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAAACAUC--- --------------((((.(...((((.....((.......)))))).--.)))))....((((((((((((((........))))..(((((((((.....))--)))))))(((.......))))))))))))).......--- ( -42.20, z-score = -1.23, R) >droSim1.chr2L 8077889 125 + 22036055 --------------GCGAGCUUUGCUAUUAUUGCACUUGUCGCGAGCC--UGUUGCCAUGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAAAAAUC--- --------------((((....(((.......)))....))))(.((.--....)))...((((((((((((((........))))..(((((((((.....))--)))))))(((.......))))))))))))).......--- ( -43.30, z-score = -0.98, R) >droSec1.super_411 5003 125 + 12313 --------------GCGACCUUUGCUAUUAUUGCACUUGUCGCGAGCC--UGUUGCCAUGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUUGGGCGAAAAAUC--- --------------(((((...(((.......)))...)))))(.((.--....)))...((((((((((((((........))))..(((((((((.....))--)))))))(((.......))))))))))))).......--- ( -44.50, z-score = -1.55, R) >droYak2.chr2L 10928026 121 + 22324452 --------------GCGAUGUUUGCUCUUAUUUCGUUUUUCGCGACCC--UGUUGCCAUGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUC------- --------------((((.((........)).)))).....(((((..--.)))))....((((((((((((((........))))..(((((((((.....))--)))))))(((.......)))))))))))))...------- ( -40.90, z-score = -1.31, R) >droEre2.scaffold_4929 17199911 120 + 26641161 --------------GCGAGCUU-ACUCUUAUUCCGUUUUUCGCGACCC--UGUUUCCAUGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUC------- --------------(((((...-((.........))..))))).....--..........((((((((((((((........))))..(((((((((.....))--)))))))(((.......)))))))))))))...------- ( -40.10, z-score = -1.39, R) >droAna3.scaffold_12916 10205954 124 - 16180835 --------------GCACGCUU---CGCUCUUAUGCUGCUUUAGCGCACGUCCAUGCAUGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCCAAC--- --------------....(((.---(((((....(((((((..(((((.((....)).)))))..)))))))..........(((((.(((((((((.....))--)))))))(((.......))))))))))))))))....--- ( -48.00, z-score = -2.06, R) >droPer1.super_1 4928339 131 - 10282868 ---------------UCGGCUCUUGUGUUCUUGUAUCGCUUAAGCGCCGUCGCUCGCGAGUUCUCGGGUGGCUGAGUUGUUAGAGCAUUGGACUGUUAACCCAAAAGGAUCCAGGGUUCAAGUCCCUGCUGGGGCGAUUCCAACUC ---------------..(((.((((.((.........)).)))).)))((((((((.(....).)))))))).((((((((..((((..(((((...(((((...........)))))..))))).))))..))))))))...... ( -46.10, z-score = -1.20, R) >dp4.chr4_group3 7817783 131 - 11692001 ---------------UCGGCUCUUGUGUUCUUGUAUCGCUUAAGCGCUGUCGCUCGCGAGUUCUCGGGUAGCUGAGUUGUUAGAGCAUUGGACUUUUAACCCAAAAGGAUCCAGGGUUCGAGUCCCUGUUGGGGCGAUUCCAACUC ---------------((((((((((.(..(((((...((....))((....))..)))))..).)))).))))))((((...((((.((((((((((......))))).))))).))))..(((((....))))).....)))).. ( -45.00, z-score = -1.41, R) >droWil1.scaffold_181089 352054 122 - 12369635 ------------------GCUCAAUUAACGUAUCGUCUGUAGGACGCGAGCAAU-GCAUAUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAUCGAUU--- ------------------((((........(((((.(((.((.((.((.(((..-.....))).)).))..))))).)))))(((((.(((((((((.....))--)))))))(((.......))))))))))))........--- ( -43.50, z-score = -2.11, R) >droVir3.scaffold_12963 16043624 120 + 20206255 -----------------GGUUU---ACUC-UUAAGCAGUCACAGCGCCUCGUUCUGCAAGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCCAUU--- -----------------(((((---....-....((((....((((...))))))))....(((((((((((((........))))..(((((((((.....))--)))))))(((.......)))))))))))))))))...--- ( -45.70, z-score = -2.74, R) >droMoj3.scaffold_6500 5488739 121 + 32352404 ----------------AGGUUU---GCUC-UUAAACAGCCACGGUGCCUCAAUUCGCAAGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCCAAC--- ----------------.(((((---((((-.......((((((.(((((.....(....).....))))).)...)).))).(((((.(((((((((.....))--)))))))(((.......)))))))))))))))))...--- ( -45.90, z-score = -2.30, R) >droGri2.scaffold_15252 13475674 138 + 17193109 GCAAAGUGCGCUCUUUUCGCUU---ACUUGUUAGUCAGCCACAGCGCCUCAGCAUGCAAGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA--GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCCAAA--- ((..((.(((((......(((.---(((....))).)))...)))))))..))..((...((((((((((((((........))))..(((((((((.....))--)))))))(((.......))))))))))))).))....--- ( -51.90, z-score = -2.16, R) >anoGam1.chr3R 13541897 124 - 53272125 -------------------CAAAGCAACAAAACUAAAGUCAUAGCUCAUGAAUCUGUGCA-GCCCGGAUAGCUCAGUCGGUAGAGCGUUGGACUUUUAAUCCAA--CGGUCUAGGGUUCAAGUCCCUAUUCGGGCGUGGUAAUGUU -------------------......((((..((((........((.((......)).)).-(((((((((..........((((.(((((((.......)))))--)).))))(((.......)))))))))))).))))..)))) ( -39.70, z-score = -2.91, R) >consensus _______________CGAGCUUUGCUCUUAUUGCACUGCUAGAGCGCC__UGUUUGCAUGUGCCCGGGUAGCUCAGUCGGUAGAGCAUUGGACUUUUAAUCCAA__GGGUCCAGGGUUCAAGUCCCUGCUCGGGCGAGCCAAC___ ............................................................((((((((((((((........))))...((((((..(((((...........))))).)))))).)))))))))).......... (-29.82 = -29.71 + -0.11)
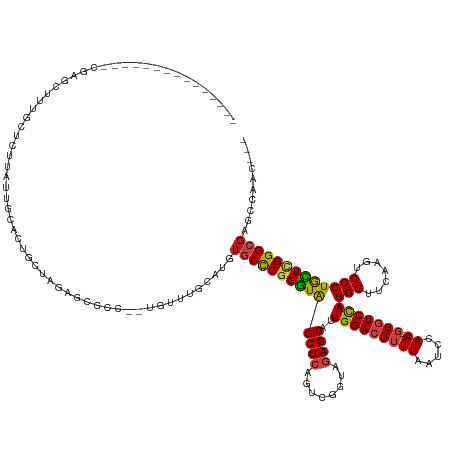
| Location | 8,290,681 – 8,290,806 |
|---|---|
| Length | 125 |
| Sequences | 13 |
| Columns | 146 |
| Reading direction | reverse |
| Mean pairwise identity | 70.93 |
| Shannon entropy | 0.60723 |
| G+C content | 0.53471 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
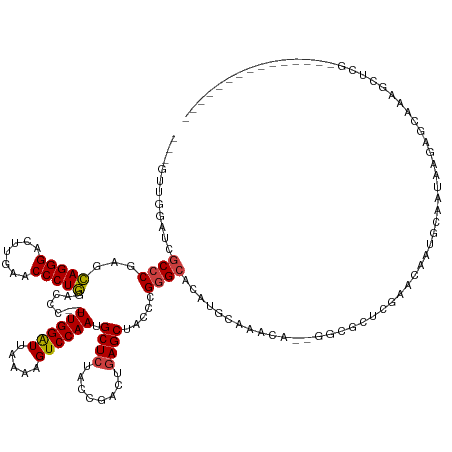
>dm3.chr2L 8290681 125 - 23011544 ---GAUGUUUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAGAUGGCAACA--GGCUCGCGAAAAAUGCAAUAAGAGCAAAGCUUGC-------------- ---..(((((((((((..(((((.......))))).....--((((((.....)))))).((((........))))....))))).)).)))).((--((((.((................))..)))))).-------------- ( -37.29, z-score = -0.88, R) >droSim1.chr2L 8077889 125 - 22036055 ---GAUUUUUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACAUGGCAACA--GGCUCGCGACAAGUGCAAUAAUAGCAAAGCUCGC-------------- ---......(((...((((((((.((........)).)))--((((((.....)))))))))))...)))..(((((.(.(((((...((....))--.))))).).....(((.......))).)))))..-------------- ( -38.70, z-score = -1.40, R) >droSec1.super_411 5003 125 - 12313 ---GAUUUUUCGCCCAAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACAUGGCAACA--GGCUCGCGACAAGUGCAAUAAUAGCAAAGGUCGC-------------- ---(((((((...((((((((((.......)))))....)--))))...)))))))....((((........)))).....((((...((....))--.))))(((((...(((.......)))...)))))-------------- ( -42.00, z-score = -2.24, R) >droYak2.chr2L 10928026 121 - 22324452 -------GAUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACAUGGCAACA--GGGUCGCGAAAAACGAAAUAAGAGCAAACAUCGC-------------- -------(((((((((..(((((.......))))).....--((((((.....)))))).((((........))))....)))))...((....))--.))))((((.....................))))-------------- ( -36.70, z-score = -1.73, R) >droEre2.scaffold_4929 17199911 120 - 26641161 -------GAUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACAUGGAAACA--GGGUCGCGAAAAACGGAAUAAGAGU-AAGCUCGC-------------- -------..((((.....(((((.......)))))(((((--...........((((...((((........)))).....))))...((....))--)))))))))............(((.-...)))..-------------- ( -38.90, z-score = -1.84, R) >droAna3.scaffold_12916 10205954 124 + 16180835 ---GUUGGCUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACAUGCAUGGACGUGCGCUAAAGCAGCAUAAGAGCG---AAGCGUGC-------------- ---(((.(((((((((..(((((.......))))).....--((((((.....)))))).((((........))))....)))))....((((....))))(((.....)))....)))).---.)))....-------------- ( -43.50, z-score = -1.40, R) >droPer1.super_1 4928339 131 + 10282868 GAGUUGGAAUCGCCCCAGCAGGGACUUGAACCCUGGAUCCUUUUGGGUUAACAGUCCAAUGCUCUAACAACUCAGCCACCCGAGAACUCGCGAGCGACGGCGCUUAAGCGAUACAAGAACACAAGAGCCGA--------------- ..(((((.......)))))..(((((..(((((...........)))))...)))))...(((((.....(((........)))...(((((((((....)))))..))))............)))))...--------------- ( -38.90, z-score = -1.30, R) >dp4.chr4_group3 7817783 131 + 11692001 GAGUUGGAAUCGCCCCAACAGGGACUCGAACCCUGGAUCCUUUUGGGUUAAAAGUCCAAUGCUCUAACAACUCAGCUACCCGAGAACUCGCGAGCGACAGCGCUUAAGCGAUACAAGAACACAAGAGCCGA--------------- ..(((((.......)))))..(((((..(((((...........)))))...)))))...(((((.....(((........)))...(((((((((....)))))..))))............)))))...--------------- ( -38.90, z-score = -2.08, R) >droWil1.scaffold_181089 352054 122 + 12369635 ---AAUCGAUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCAUAUGC-AUUGCUCGCGUCCUACAGACGAUACGUUAAUUGAGC------------------ ---........(((((..(((((.......))))).....--((((((.....)))))).((((........))))....)))))......-...(((((((((.....))))..........)))))------------------ ( -35.40, z-score = -1.59, R) >droVir3.scaffold_12963 16043624 120 - 20206255 ---AAUGGCUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACUUGCAGAACGAGGCGCUGUGACUGCUUAA-GAGU---AAACC----------------- ---....(((((((((..(((((.......))))).....--((((((.....)))))).((((........))))....)))))....((((.(((......)))..))))....-))))---.....----------------- ( -39.70, z-score = -2.13, R) >droMoj3.scaffold_6500 5488739 121 - 32352404 ---GUUGGCUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACUUGCGAAUUGAGGCACCGUGGCUGUUUAA-GAGC---AAACCU---------------- ---(((.((((....((((((((.((........)).)))--((((((.....))))))))))).......(((((..(((((((.(((.......))))).))).))..))))).-))))---.)))..---------------- ( -39.90, z-score = -1.32, R) >droGri2.scaffold_15252 13475674 138 - 17193109 ---UUUGGCUCGCCCGAGCAGGGACUUGAACCCUGGACCC--UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACUUGCAUGCUGAGGCGCUGUGGCUGACUAACAAGU---AAGCGAAAAGAGCGCACUUUGC ---....((..(((((..(((((.......))))).....--((((((.....)))))).((((........))))....)))))....))..((.((((((((.(.(((.(((....)))---.)))....).))))).))).)) ( -46.90, z-score = -1.42, R) >anoGam1.chr3R 13541897 124 + 53272125 AACAUUACCACGCCCGAAUAGGGACUUGAACCCUAGACCG--UUGGAUUAAAAGUCCAACGCUCUACCGACUGAGCUAUCCGGGC-UGCACAGAUUCAUGAGCUAUGACUUUAGUUUUGUUGCUUUG------------------- ...........(((((..(((((.......)))))....(--((((((.....)))))))((((........))))....)))))-.(((((((.....(((......)))....)))).)))....------------------- ( -36.50, z-score = -3.31, R) >consensus ___GUUGGAUCGCCCGAGCAGGGACUUGAACCCUGGACCC__UUGGAUUAAAAGUCCAAUGCUCUACCGACUGAGCUACCCGGGCACAUGCAAACA__GGCGCUCGAACAAUGCAAUAAGAGCAAAGCUCG_______________ ...........((((...(((((.......))))).......((((((.....)))))).((((........)))).....))))............................................................. (-23.49 = -23.68 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:39 2011