
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,151,910 – 18,152,055 |
| Length | 145 |
| Max. P | 0.530460 |

| Location | 18,151,910 – 18,152,055 |
|---|---|
| Length | 145 |
| Sequences | 8 |
| Columns | 163 |
| Reading direction | reverse |
| Mean pairwise identity | 79.03 |
| Shannon entropy | 0.41471 |
| G+C content | 0.51095 |
| Mean single sequence MFE | -47.79 |
| Consensus MFE | -24.11 |
| Energy contribution | -25.48 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
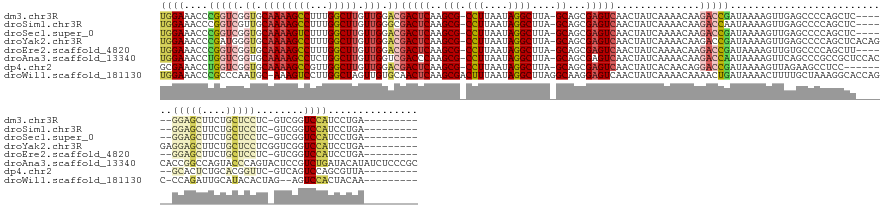
>dm3.chr3R 18151910 145 - 27905053 UGGAAACCCGGUCGGUGCAAAAGCCUUUGGCUUGUUGGACGACUCAAGCG-CCUUAAUAGGCUUA-GCAGCGAGUCAACUAUCAAAACAAGACCGAUAAAAGUUGAGCCCCAGCUC------GGAGCUUCUGCUCCUC-GUCGGUCCAUCCUGA--------- .(((.....(((.(((.((((((((...))))).))).))(((((..(((-(((....))))...-))...)))))..).))).......(((((((.......((((....))))------(((((....)))))..-)))))))..)))...--------- ( -51.00, z-score = -2.26, R) >droSim1.chr3R 17958744 145 - 27517382 UGGAAACCCGGUCGUUGCAAAAGCCUUUGGCUUGUUGGGCGACUCAAGCG-CCUUAAUAGGCUUA-GCAGCGAGUCAACUAUCAAAACAAGACCAAUAAAAGUUGAGCCCCAGCUC------GGAGCUUCUGCUCCUC-GUCGGUCCAUCCUGA--------- .(((.....(((((((((..((((((..(((...(((((...)))))..)-)).....)))))).-))))))).))..............((((..........((((....))))------(((((....)))))..-...))))..)))...--------- ( -48.00, z-score = -1.58, R) >droSec1.super_0 18507121 145 - 21120651 UGGAAACCCGGUCGGUGCAAAAGUCUUUGGCUUGUUGGACGACUCAAGCG-CCUUAAUAGGCUUA-GCAGCGAGUCAACUAUCAAAACAAGACCGAUAAAAGUUGAGCCCCAGCUC------GGAGCUUCUGCUCCUC-GUCGGUCCAUCCUGA--------- .(((.....(((((.(((...........(((((..(.....).)))))(-(((....))))...-))).))).))..............(((((((.......((((....))))------(((((....)))))..-)))))))..)))...--------- ( -49.00, z-score = -1.88, R) >droYak2.chr3R 18995182 152 - 28832112 UGGAAACCCGAUGGGUGCAAAAGCCUUUGGCUUGUUGGACGACUCAAGCG-CCUUAAUAGGCUUA-GCAGCGAGUCAACUAUCAAAACAAGACCGAUAAAAGUUGAGCCCCAGCUCACAGGAGGAGCUUCUGCUCCUCGGUCGGUCCAUCCUGA--------- .(((.....(((((((.((((((((...))))).))).))(((((..(((-(((....))))...-))...)))))..))))).......(((((((....((.((((....))))))..(((((((....))))))).)))))))..)))...--------- ( -61.30, z-score = -3.91, R) >droEre2.scaffold_4820 540605 145 + 10470090 UGGAAACCCGGUCGGUGCAAAAGCCUUUGGCUUGUUGGACGACUCAAGCG-CCUUAAUAGGCUUA-GCAGCGAGUCAACUAUCAAAACAAGACCGAUAAAAGUUGUGCCCCAGCUU------GGAGCUUCUGCUCCUC-GUCGGUCCAUCCUGA--------- .(((.....(((.(((.((((((((...))))).))).))(((((..(((-(((....))))...-))...)))))..).))).......(((((((..((((((.....))))))------(((((....)))))..-)))))))..)))...--------- ( -49.80, z-score = -2.00, R) >droAna3.scaffold_13340 9133649 161 - 23697760 UGGAAACCUGGUCGGUGCAAAAGCCUCUGGCUUGUUGGUCGACCCAAGCG-CCUUAAUAGGCUUA-GCAGCGAGUCAACUAUCAAAACAAGACCAAUAAAAGUUCAGCCCGCCGCUCCACCACCGGCCAGUACCCAGUACUCCGUCUGAUACAUAUCUCCCGC .(((...((((((((((....(((..(.((((((((((((.......(((-(((....))))...-))...((........)).......)))))))).......)))).)..)))....))))))))))....(((........))).........)))... ( -46.71, z-score = -1.85, R) >dp4.chr2 12306091 143 - 30794189 GCGAAACCUGGUCGGUGCAAAAGCCGUUGGCUUGUUGGACGACUCAAGCG-CCUUAAUAGGCUUA-GCAGCGAGUCAACUAUCACAACAGGACCGAUAAAAGUUAGAAGCCUCC--------GCACUCUGCACGGUUC-GUCAGUCCAGCGUUA--------- ....(((((((.((((......)))((((((((((((((....))....(-(((....))))...-.))))))))))))..........((((((......((.....))....--------((.....)).))))))-....).)))).))).--------- ( -42.90, z-score = -0.22, R) >droWil1.scaffold_181130 12297332 150 - 16660200 UGGAAACCCGCCCAAUGC-AAAGUCCUUGGCUAGUUGUGCAACUCAAGCGACUUUAAUAGGCUUAGGCAAGGAGUCAACUAUCAAAACAAAACUGAUAAAACUUUUGCUAAAGGCACCAGC-CCAGAUUGCAUACACUAG--AGUCCACUACAA--------- ((((.......((((.(.-.....).))))((((((((((((.((.(((((((((.....((....))..))))))...(((((.........)))))........)))...(((....))-)..))))))))).)))))--..))))......--------- ( -33.60, z-score = -0.10, R) >consensus UGGAAACCCGGUCGGUGCAAAAGCCUUUGGCUUGUUGGACGACUCAAGCG_CCUUAAUAGGCUUA_GCAGCGAGUCAACUAUCAAAACAAGACCGAUAAAAGUUGAGCCCCAGCUC______GGAGCUUCUGCUCCUC_GUCGGUCCAUCCUGA_________ ((((....(((((.((.((((((((...))))).))).))(((((..((..(((....))).....))...)))))..............)))))...........................(((((....)))))........))))............... (-24.11 = -25.48 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:01 2011