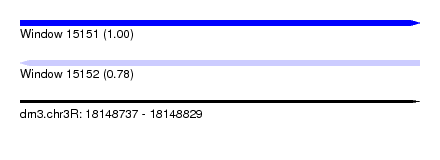
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,148,737 – 18,148,829 |
| Length | 92 |
| Max. P | 0.996784 |

| Location | 18,148,737 – 18,148,829 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.42 |
| Shannon entropy | 0.57213 |
| G+C content | 0.47553 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -12.47 |
| Energy contribution | -14.94 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
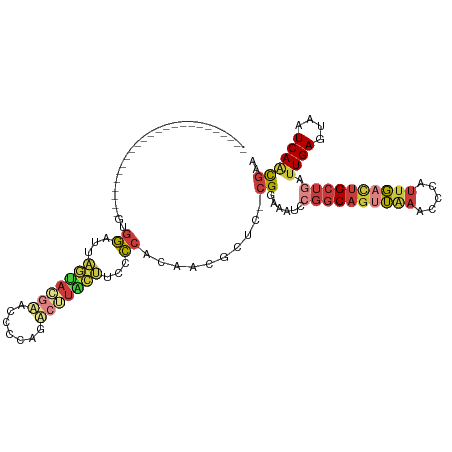
>dm3.chr3R 18148737 92 + 27905053 ------------------------GUGGAUUAGUACGAACCCCAGUCGUACUUCCCCACAACGCUC-CGGAAAUCCGGCAGUUAAACCCAUUGACUGCUGAUUGAGUAAUCAACGAA ------------------------((((...(((((((.......)))))))...))))...((((-(((....)))((((((((.....)))))))).....)))).......... ( -30.70, z-score = -4.53, R) >droSim1.chr3R 17955710 92 + 27517382 ------------------------AUGGAUUAGUACGAACCCCAGUCGUACUUCCCCACAACGCUC-CGGAAAUCCGGCAGUUAAACCCAUUGACUGCUGAUUGAGUAAUCAACGAA ------------------------.(((...(((((((.......)))))))...)))....((((-(((....)))((((((((.....)))))))).....)))).......... ( -27.10, z-score = -3.57, R) >droSec1.super_0 18504058 92 + 21120651 ------------------------AUGGAUUAGUACGAACCCCAGUCGUACUUCCCCACAUCGCUC-CGGAAAUCCGGCAGUUAAACCCAUUGACUGCUGAUUGAGUAAUCAACGAA ------------------------.(((...(((((((.......)))))))...)))..(((...-........((((((((((.....)))))))))).((((....))))))). ( -27.70, z-score = -3.64, R) >droYak2.chr3R 18991976 92 + 28832112 ------------------------GUGGAUUAGUAAGAGCCCCAGACUUGCUUCCCCACAACGCUG-CGGAAAUCCGGCAGUUAAACCCAUUGACUGCUGAUUGAGUAAUCAACGAA ------------------------((((...((((((.........))))))...)))).......-((......((((((((((.....)))))))))).((((....)))))).. ( -26.80, z-score = -2.84, R) >droEre2.scaffold_4820 537521 92 - 10470090 ------------------------GUGGAUUAGUGUGAGCCCCAAACUAACUUCCCCACAAUGCUG-CGGAAAUCCGGCAGUUAAACCCAUUGACUGCUGAUUGAGUAAUCAACGAA ------------------------((((.(((((.((.....)).))))).....)))).......-((......((((((((((.....)))))))))).((((....)))))).. ( -23.60, z-score = -2.15, R) >droAna3.scaffold_13340 9130581 96 + 23697760 -------------------CGGAUACGGAUAUGCAAUUUCCGCAAACUUUGGGCCCCCCCAUGUUUUCGAAAAUCAGGCA--UGAACCCAUUGGCUGCUGAUUGAGUAAUCAGCGAA -------------------.......((.(((((.((((.((.((((..((((....)))).)))).)).))))...)))--))...))......((((((((....)))))))).. ( -24.50, z-score = -0.23, R) >dp4.chr2 12302758 114 + 30794189 GUGGCUUUAAGAGUGCUCUCAGGGGUCUCACCUCACAUACCCCCGAAUUGACCCUAAUCCAUGAUUAUGAACCCCAUUCAGCGAUAUUC---UAUCGUUGAUUGAAUAAUCACUCAG ((((......(((....))).((((((......................))))))...))))((((((......((.(((((((((...---))))))))).)).))))))...... ( -27.35, z-score = -1.83, R) >consensus ________________________GUGGAUUAGUACGAACCCCAGACUUACUUCCCCACAACGCUC_CGGAAAUCCGGCAGUUAAACCCAUUGACUGCUGAUUGAGUAAUCAACGAA ..........................((...(((((((.......)))))))...))..................((((((((((.....)))))))))).((((....)))).... (-12.47 = -14.94 + 2.48)

| Location | 18,148,737 – 18,148,829 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.42 |
| Shannon entropy | 0.57213 |
| G+C content | 0.47553 |
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.99 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18148737 92 - 27905053 UUCGUUGAUUACUCAAUCAGCAGUCAAUGGGUUUAACUGCCGGAUUUCCG-GAGCGUUGUGGGGAAGUACGACUGGGGUUCGUACUAAUCCAC------------------------ ((((((((((.((.....)).))))))))))...(((..((((....)))-)...)))(((((..((((((((....).)))))))..)))))------------------------ ( -30.40, z-score = -2.17, R) >droSim1.chr3R 17955710 92 - 27517382 UUCGUUGAUUACUCAAUCAGCAGUCAAUGGGUUUAACUGCCGGAUUUCCG-GAGCGUUGUGGGGAAGUACGACUGGGGUUCGUACUAAUCCAU------------------------ ((((((((((.((.....)).))))))))))..((((..((((....)))-)...))))((((..((((((((....).)))))))..)))).------------------------ ( -29.40, z-score = -1.86, R) >droSec1.super_0 18504058 92 - 21120651 UUCGUUGAUUACUCAAUCAGCAGUCAAUGGGUUUAACUGCCGGAUUUCCG-GAGCGAUGUGGGGAAGUACGACUGGGGUUCGUACUAAUCCAU------------------------ ((((((((((.((.....)).)))))))))).....((.((((....)))-)))....(((((..((((((((....).)))))))..)))))------------------------ ( -28.60, z-score = -1.55, R) >droYak2.chr3R 18991976 92 - 28832112 UUCGUUGAUUACUCAAUCAGCAGUCAAUGGGUUUAACUGCCGGAUUUCCG-CAGCGUUGUGGGGAAGCAAGUCUGGGGCUCUUACUAAUCCAC------------------------ ...(((((((....)))))))......(((((((((..(((((....)).-(((..((((......))))..))).)))..)))..)))))).------------------------ ( -23.70, z-score = -0.19, R) >droEre2.scaffold_4820 537521 92 + 10470090 UUCGUUGAUUACUCAAUCAGCAGUCAAUGGGUUUAACUGCCGGAUUUCCG-CAGCAUUGUGGGGAAGUUAGUUUGGGGCUCACACUAAUCCAC------------------------ ..((((((((.((.....)).))))))))(((......)))((((..(((-(......))))..)..(((((.((.....)).))))))))..------------------------ ( -24.00, z-score = -0.84, R) >droAna3.scaffold_13340 9130581 96 - 23697760 UUCGCUGAUUACUCAAUCAGCAGCCAAUGGGUUCA--UGCCUGAUUUUCGAAAACAUGGGGGGGCCCAAAGUUUGCGGAAAUUGCAUAUCCGUAUCCG------------------- ...(((((((....))))))).....((((((..(--(((..((((((((.((((.((((....))))..)))).)))))))))))))))))).....------------------- ( -31.30, z-score = -1.87, R) >dp4.chr2 12302758 114 - 30794189 CUGAGUGAUUAUUCAAUCAACGAUA---GAAUAUCGCUGAAUGGGGUUCAUAAUCAUGGAUUAGGGUCAAUUCGGGGGUAUGUGAGGUGAGACCCCUGAGAGCACUCUUAAAGCCAC .(((((((...((((.(((.(((((---...))))).))).))))..))))..)))(((...(((((...(((((((((............)))))))))...))))).....))). ( -32.20, z-score = -1.02, R) >consensus UUCGUUGAUUACUCAAUCAGCAGUCAAUGGGUUUAACUGCCGGAUUUCCG_GAGCGUUGUGGGGAAGUAAGUCUGGGGUUCGUACUAAUCCAC________________________ ...(((((((....)))))))................((((((....)))...)))..(((((..(((((((.......)))))))..)))))........................ (-13.32 = -13.99 + 0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:35:00 2011