
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,131,642 – 18,131,779 |
| Length | 137 |
| Max. P | 0.999218 |

| Location | 18,131,642 – 18,131,751 |
|---|---|
| Length | 109 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Shannon entropy | 0.44337 |
| G+C content | 0.51524 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -22.24 |
| Energy contribution | -21.64 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18131642 109 - 27905053 ---UGUCGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUUCUUGCGCCCAAUUCCACCCACUUUGCCGCAU ---..((((((.(((((..-.)))))))))))(((((((((((......)))))))))))........(((((((..-(((.....))).................))))))). ( -32.31, z-score = -1.88, R) >droPer1.super_0 3638689 95 + 11822988 ------AGUCGUCGUCGUC-UGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUACUUGC-CGCACUUCGGCUCCU---------- ------..((((((.(((.-...)))))))))(((((((((((......))))))))))).........(((((...-.)))))..((-((.....))))....---------- ( -30.80, z-score = -1.58, R) >dp4.chr2 12286650 101 - 30794189 AGUCGUCGUCGUCGUCGUC-UGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUACUUGC-CGCACUUCGGCUCCG---------- (((((((((((.((((...-..))))))))))(((((((((((......)))))))))))........(((((((..-.......)))-))))...)))))...---------- ( -33.80, z-score = -1.76, R) >droAna3.scaffold_13340 9113676 98 - 23697760 --UUUCUGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUACUUGCUGCCAAUCCCACCC------------ --...((((((.(((((..-.)))))))))))(((((((((((......)))))))))))..........(((((..-.((.....))))))).........------------ ( -28.70, z-score = -2.43, R) >droYak2.chr3R 18973530 109 - 28832112 ---UGUCGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUUCUUGCGCCCAAUGCCACCCACUUUGCGGCAU ---(((((((((((.(((.-...)))))))).(((((((((((......))))))))))).........((((((..-(((.....))).....)))))).......)))))). ( -37.00, z-score = -2.14, R) >droSec1.super_0 18487141 109 - 21120651 ---UGUCCUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC-CGCUUCUUGCGCCCAAUUCCACCCACUUUGCCGCAU ---.....((((((.(((.-...)))))))))(((((((((((......)))))))))))........(((((((..-(((.....))).................))))))). ( -30.11, z-score = -1.67, R) >droSim1.chr3R 17937023 109 - 27517382 ---UGUCGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUAUUGUGGCACC-CGCUUCUUGCGCCCAAUUCCACCCACUUUGCCGCAU ---..((((((.(((((..-.)))))))))))(((((((((((......)))))))))))........(((((((..-(((.....))).................))))))). ( -32.31, z-score = -2.28, R) >droVir3.scaffold_13047 10872021 99 + 19223366 ---CCUUGUUGUUGUCCUCCAGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCACC-CACAGCAACAGCCAACGUGCAACUC----------- ---....(((((.((((....))))(((.((((((((((((((......))))))))))......((((((((....-..)))))))))))).))))))))..----------- ( -34.90, z-score = -3.26, R) >droMoj3.scaffold_6540 27824024 98 - 34148556 ---CCUUGUUGUUGUCCUCGAGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCACC-CACAGCCA-GGCAAACGUGUAACUC----------- ---(((.(((((.((((....))))((((((((((((((((((......)))))))))).)))))))).........-.))))).)-))..............----------- ( -29.20, z-score = -1.38, R) >droGri2.scaffold_15074 1267681 99 - 7742996 ---CCUUGUUGUUGUCCUCCUGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCAUCGCGCAGCAA-GGCGAGCGUGCAUCGC----------- ---(((((((((.((((....))))((((((.(((((((((((......)))))))))))..((....))...)))))))))))))-))...(((.....)))----------- ( -37.50, z-score = -2.10, R) >consensus ___UGUCGUCGUCGUCGUC_UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC_CGCUUCUUGCGCCCAAUUCCACCCC___________ .......((((.(((((....)))))))))(((((((((((((......))))))))(((((....))))).)))))..................................... (-22.24 = -21.64 + -0.60)


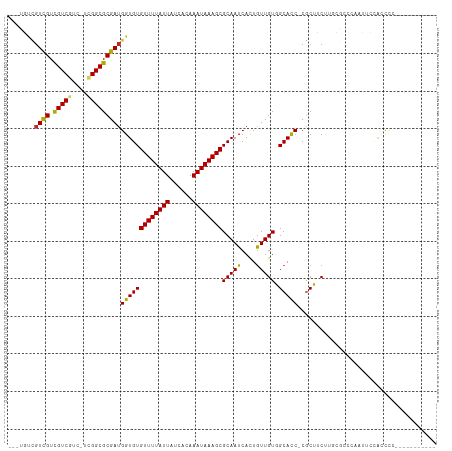
| Location | 18,131,678 – 18,131,779 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.11 |
| Shannon entropy | 0.48383 |
| G+C content | 0.50625 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -22.04 |
| Energy contribution | -21.44 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
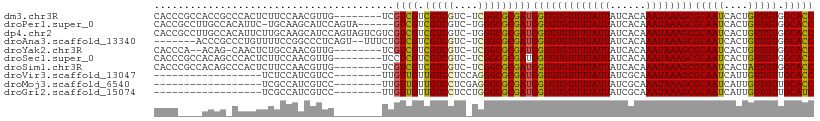
>dm3.chr3R 18131678 101 - 27905053 CACCCGCCACCGCCCACUCUUCCAACGUUG--------UCGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC .....(((((.((.................--------((((((.(((((..-.)))))))))))(((((((((((......)))))))))))......)).)))))... ( -32.50, z-score = -2.94, R) >droPer1.super_0 3638714 102 + 11822988 CACCGCCUUGCCACAUUC-UGCAAGCAUCCAGUA------GUCGUCGUCGUC-UGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC ........(((((((...-((....))..((((.------.((((((.(((.-...)))))))))(((((((((((......)))))))))))...)))).))))))).. ( -33.20, z-score = -2.15, R) >dp4.chr2 12286675 109 - 30794189 CACCGCCUUGCCACAUUCUUGCAAGCAUCCAGUAGUCGUCGUCGUCGUCGUC-UGGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC ........(((((((.........((..((((....((.((....)).)).)-)))..))(((..(((((((((((......)))))))))))))).....))))))).. ( -35.00, z-score = -2.30, R) >droAna3.scaffold_13340 9113700 100 - 23697760 -------ACCCGCCCUGUUUUCCGGCCCUCAGU--UUUCUGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC -------.................((((.((((--...((((((.(((((..-.)))))))))))(((((((((((......)))))))))))...))))..).)))... ( -29.60, z-score = -2.44, R) >droYak2.chr3R 18973566 98 - 28832112 CACCCA--ACAG-CAACUCUGCCAACGUUG--------UCGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC ..((((--((((-......(((..(((..(--------(((.(((((.....-.)))))))))..)))((((((((......)))))))))))....)))))).)).... ( -31.90, z-score = -2.68, R) >droSec1.super_0 18487177 101 - 21120651 CACCCGCCACAGCCCACUCUUCCAACGUUG--------UCCUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC .....((((((((.................--------...((((((.(((.-...)))))))))(((((((((((......)))))))))))......))))))))... ( -34.10, z-score = -4.05, R) >droSim1.chr3R 17937059 101 - 27517382 CACCCGCCACAGCCCACUCUUCCAACGUUG--------UCGUCGUCGUCGUC-UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUAUUGUGGCACC .....(((((((..................--------((((((.(((((..-.)))))))))))(((((((((((......))))))))))).......)))))))... ( -33.30, z-score = -3.81, R) >droVir3.scaffold_13047 10872046 84 + 19223366 ------------------UCUCCAUCGUCC--------UUGUUGUUGUCCUCCAGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCACC ------------------...(((((((((--------(((..(.....)..))))).)))))))(((((((((((......)))))))))))................. ( -25.40, z-score = -3.16, R) >droMoj3.scaffold_6540 27824048 84 - 34148556 ------------------UCGCCAUCGUCC--------UUGUUGUUGUCCUCGAGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCACC ------------------.(((((((((((--------(((..(....)..)))))..))))))))).((((((((......))))))))((((.((....))))))... ( -28.30, z-score = -3.06, R) >droGri2.scaffold_15074 1267706 84 - 7742996 ------------------UCGCCAUCGUCC--------UUGUUGUUGUCCUCCUGGGCGCGAUGGUGUGUUUUAUUAUCGCAAAUAAAGCGCAAUCAUUGUUGUUGCAUC ------------------.(((((((((..--------........((((....))))))))))))).((((((((......))))))))((((.((....))))))... ( -26.10, z-score = -3.05, R) >consensus CACC____ACCG_CCACUUUGCCAACGUCC________UCGUCGUCGUCGUC_UCGGCGCGAUGGUGUGUUUUAUUAUCACAAAUAAAGCGCAAUCACUGUUGUGGCACC ........................................((((.(((((....)))))))))(((((((((((((......))))))))(((((....))))).))))) (-22.04 = -21.44 + -0.60)
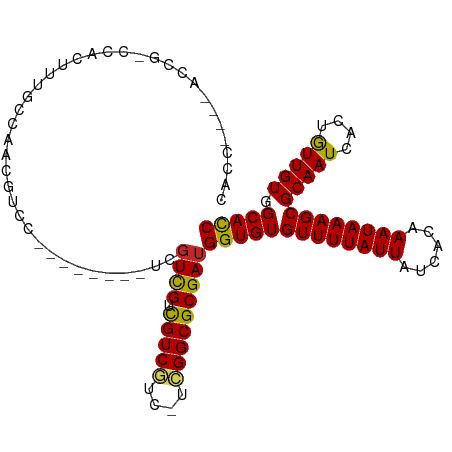

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:54 2011