| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,119,861 – 18,119,972 |
| Length | 111 |
| Max. P | 0.519361 |

| Location | 18,119,861 – 18,119,972 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Shannon entropy | 0.45422 |
| G+C content | 0.55200 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -18.55 |
| Energy contribution | -22.41 |
| Covariance contribution | 3.86 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
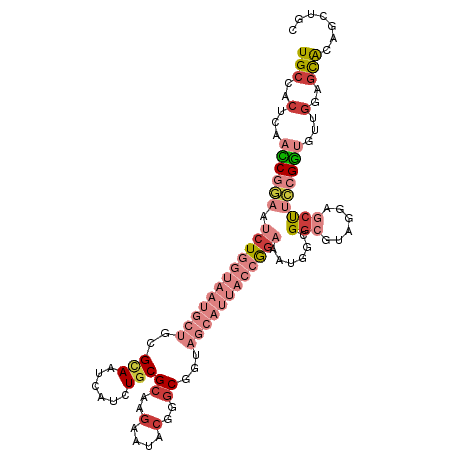
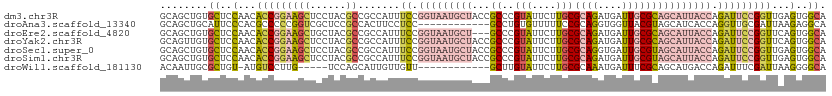
>dm3.chr3R 18119861 111 + 27905053 UGCCACUCAACCGGAAUCUGGUAAUGCUGCGCAAUCAUCUGCGCAAGAAUACGGGCGGUAGCAUUACCGGAAAUGGCGGCGUAGGAGCUUCCGGUGUUGGAGCACAGCUGC (((..(...((((((.(((((((((((((((((......)))((..(....)..)).))))))))))))))......(((......)))))))))...)..)))....... ( -44.70, z-score = -2.16, R) >droAna3.scaffold_13340 9104064 99 + 23697760 UGCCUCUUAAUCGCAACCUGGUGAUGCUACGUAACCACCUGCGGAAAAACACAGGC------------GGAGGAAGUGGCGGAGCGACCGGGGGCGUGGGAAUGCAGCUGC (((.(((((..(((..(((((((.((((((....((.((((.(......).)))).------------)).....))))))...).)))))).))))))))..)))..... ( -35.90, z-score = -0.86, R) >droEre2.scaffold_4820 510458 108 - 10470090 UGCCACUGAACCGGAAUCUGGUAAUGCUGCGCAAUCAUCUGCGCAAGAAUACGGGC---AGCAUUACCGGAAAUGGCGGCGUAGCAGCUUCCGGUGUUGGAGCACAGCUGC (((..(...((((((.((((((((((((((.(.....(((.....)))....).))---))))))))))))...(((.((...)).)))))))))...)..)))....... ( -44.20, z-score = -2.25, R) >droYak2.chr3R 18963585 111 + 28832112 UGCCACUGAACCGGAAUCUGGUAAUGCUGCGCAAUCAUCUGCGCAAGAAUACGGGCGGUAGCAUUACCGGAAAUGGCGGCGUAGGAGCUUCCGGUGUUGGAGCACAACUGC (((..(...((((((.(((((((((((((((((......)))((..(....)..)).))))))))))))))......(((......)))))))))...)..)))....... ( -44.70, z-score = -2.58, R) >droSec1.super_0 18477394 111 + 21120651 UGCCACUCAACCGGAAUCUGGUAAUGCUACGCAAUCACCUGCGCAAGAAUACGGGCGGUAGCAUUACCGGAAAUGGCGGCGUAGGAGCUUCCGGUGUUGGAGCACAGCUGC (((..(...((((((.(((((((((((((((((......)))((..(....)..)).))))))))))))))......(((......)))))))))...)..)))....... ( -44.70, z-score = -2.46, R) >droSim1.chr3R 17927237 111 + 27517382 UGCCACUCAACCGGAAUCUGGUAAUGCUACGCAAUCAUCUGCGCAAGAAUACGGGCGGUAGCAUUACCGGAAAUGGCGGCGUAGGAGCUUCCGGUGUUGGAGCACAGCUGC (((..(...((((((.(((((((((((((((((......)))((..(....)..)).))))))))))))))......(((......)))))))))...)..)))....... ( -44.70, z-score = -2.65, R) >droWil1.scaffold_181130 12260812 93 + 16660200 UGCCCCUUAAUCGAAAUCUGGUCAUGCUGCGAAAUCAUUUGCGCAAGAAUACAAGC------------AACAACAAUGCUGGA-----CAAGGACAU-ACAGCGCAAUUGU (((...............((.((.(((.(((((....)))))))).))...))...------------.........((((..-----.........-.)))))))..... ( -15.70, z-score = 1.40, R) >consensus UGCCACUCAACCGGAAUCUGGUAAUGCUGCGCAAUCAUCUGCGCAAGAAUACGGGCGGUAGCAUUACCGGAAAUGGCGGCGUAGGAGCUUCCGGUGUUGGAGCACAGCUGC (((..(...((((((.((((((((((((..(((......)))((..(....)..))...))))))))))))......(((......)))))))))...)..)))....... (-18.55 = -22.41 + 3.86)


| Location | 18,119,861 – 18,119,972 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Shannon entropy | 0.45422 |
| G+C content | 0.55200 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
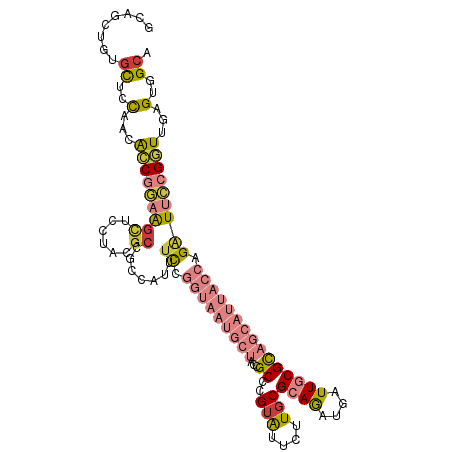
>dm3.chr3R 18119861 111 - 27905053 GCAGCUGUGCUCCAACACCGGAAGCUCCUACGCCGCCAUUUCCGGUAAUGCUACCGCCCGUAUUCUUGCGCAGAUGAUUGCGCAGCAUUACCAGAUUCCGGUUGAGUGGCA ...(((..((((....(((((((((......)).......((.(((((((((((.....)))....(((((((....))))))))))))))).))))))))).))))))). ( -38.60, z-score = -1.84, R) >droAna3.scaffold_13340 9104064 99 - 23697760 GCAGCUGCAUUCCCACGCCCCCGGUCGCUCCGCCACUUCCUCC------------GCCUGUGUUUUUCCGCAGGUGGUUACGUAGCAUCACCAGGUUGCGAUUAAGAGGCA ((....))........((((..((((((.((((.((.....((------------(((((((......)))))))))....)).)).......))..))))))..).))). ( -31.71, z-score = -1.29, R) >droEre2.scaffold_4820 510458 108 + 10470090 GCAGCUGUGCUCCAACACCGGAAGCUGCUACGCCGCCAUUUCCGGUAAUGCU---GCCCGUAUUCUUGCGCAGAUGAUUGCGCAGCAUUACCAGAUUCCGGUUCAGUGGCA ((((((....(((......)))))))))...(((((.....(((((((((((---((..(((....)))((((....))))))))))))))).(....)))....))))). ( -44.10, z-score = -3.10, R) >droYak2.chr3R 18963585 111 - 28832112 GCAGUUGUGCUCCAACACCGGAAGCUCCUACGCCGCCAUUUCCGGUAAUGCUACCGCCCGUAUUCUUGCGCAGAUGAUUGCGCAGCAUUACCAGAUUCCGGUUCAGUGGCA ((.((((.....))))(((((((((......)).......((.(((((((((((.....)))....(((((((....))))))))))))))).)))))))))......)). ( -36.60, z-score = -1.74, R) >droSec1.super_0 18477394 111 - 21120651 GCAGCUGUGCUCCAACACCGGAAGCUCCUACGCCGCCAUUUCCGGUAAUGCUACCGCCCGUAUUCUUGCGCAGGUGAUUGCGUAGCAUUACCAGAUUCCGGUUGAGUGGCA ...(((..((((....(((((((((......)).......((.(((((((((((((((((((....))))..)))).....))))))))))).))))))))).))))))). ( -41.50, z-score = -2.37, R) >droSim1.chr3R 17927237 111 - 27517382 GCAGCUGUGCUCCAACACCGGAAGCUCCUACGCCGCCAUUUCCGGUAAUGCUACCGCCCGUAUUCUUGCGCAGAUGAUUGCGUAGCAUUACCAGAUUCCGGUUGAGUGGCA ...(((..((((....(((((((((......)).......((.(((((((((((.....(((....)))((((....))))))))))))))).))))))))).))))))). ( -39.50, z-score = -2.34, R) >droWil1.scaffold_181130 12260812 93 - 16660200 ACAAUUGCGCUGU-AUGUCCUUG-----UCCAGCAUUGUUGUU------------GCUUGUAUUCUUGCGCAAAUGAUUUCGCAGCAUGACCAGAUUUCGAUUAAGGGGCA .....((((((((-..(((.(((-----(.(((((....))))------------)...(((....)))))))..)))...)))))....((...((......)).))))) ( -17.40, z-score = 2.18, R) >consensus GCAGCUGUGCUCCAACACCGGAAGCUCCUACGCCGCCAUUUCCGGUAAUGCUACCGCCCGUAUUCUUGCGCAGAUGAUUGCGCAGCAUUACCAGAUUCCGGUUGAGUGGCA ........((..(...(((((((...................(((((....)))))...(((...((((((((....))))))))...)))....)))))))...)..)). (-17.91 = -18.53 + 0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:52 2011