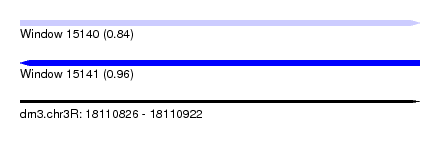
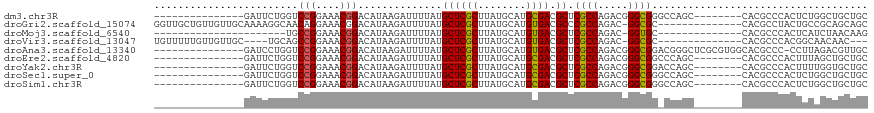
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,110,826 – 18,110,922 |
| Length | 96 |
| Max. P | 0.956212 |

| Location | 18,110,826 – 18,110,922 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Shannon entropy | 0.45333 |
| G+C content | 0.57654 |
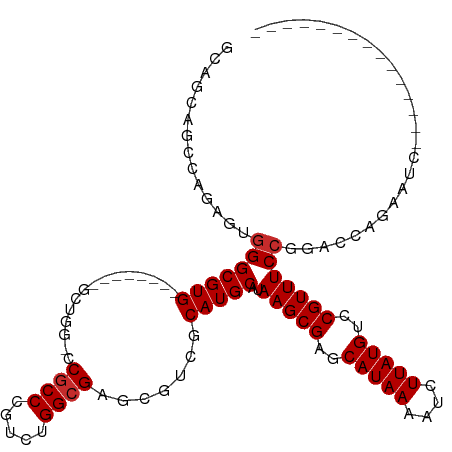
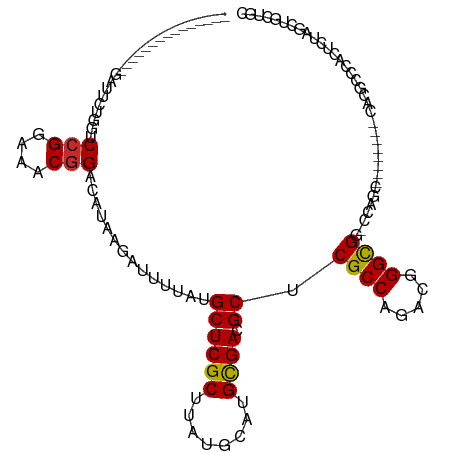
| Mean single sequence MFE | -38.11 |
| Consensus MFE | -17.43 |
| Energy contribution | -17.76 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.844199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
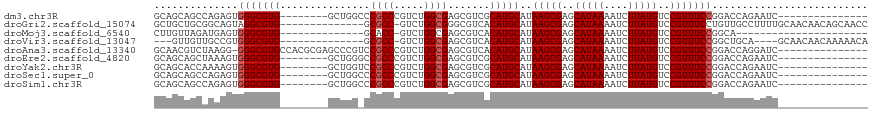
>dm3.chr3R 18110826 96 + 27905053 GCAGCAGCCAGAGUGGGCGUG--------GCUGGCCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGACCAGAAUC--------------- ((.((.((((((.((((((.(--------(....))))))))))))))..)).((((........))))))......(((...(((((....))))).)))...--------------- ( -43.00, z-score = -3.28, R) >droGri2.scaffold_15074 1246371 104 + 7742996 GCUGCUGCGGCAGUAGGCGUG--------------GCGCC-GUCUGGCGGGCGUCACAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCUGUUGCCUUUUGCAACAACAGCAACC ((((.((..((((.(((((((--------------(((((-........))))))))..(((.(((((..(((((....)))))..)))))..))).)))).))))..)).)))).... ( -46.10, z-score = -3.42, R) >droMoj3.scaffold_6540 27803042 82 + 34148556 CUUGUUAGAUGAGUGGGCGUG--------------GCACC-GUCUGGCGAGCGUCACAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGCA---------------------- (((((((((((.(((......--------------.))))-)))))))))).......(((..(((((..(((((....)))))..)))))...)))---------------------- ( -27.90, z-score = -1.34, R) >droVir3.scaffold_13047 10851089 97 - 19223366 ---GUUGUUGCCGUGGGCGUG--------------GCGCC-GUCUGGCGAGCGUCACAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGCUGCA----GCAACAACAAAAACA ---((((((((.(..(.((((--------------(((((-(.....)).)))))))......(((((..(((((....)))))..)))))..).)..).----))))))))....... ( -42.10, z-score = -3.59, R) >droAna3.scaffold_13340 9095573 103 + 23697760 GCAACGUCUAAGG-GGGCGUGCCACGCGAGCCCGUCCGCCCGUCUGGCGAGCGUCACAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGACCAGGAUC--------------- .....((((....-(((((..(..(....)...)..)))))((((((...((((...))))..(((((..(((((....)))))..)))))))))))..)))).--------------- ( -33.30, z-score = 0.04, R) >droEre2.scaffold_4820 500101 96 - 10470090 GCAGCAGCUAAAGUGGGCGUG--------GCUGGGCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGACCAGAAUC--------------- ((.((.((((...(((((..(--------((...))))))))..))))..)).((((........))))))......(((...(((((....))))).)))...--------------- ( -31.10, z-score = 0.39, R) >droYak2.chr3R 18954236 96 + 28832112 GCAGCACCAAAAGUGGGCGUG--------GCUGGUCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGACCAGAAUC--------------- ...((.(((....)))))...--------.((((((((...((...(((.....)))..))..(((((..(((((....)))))..))))).))))))))....--------------- ( -33.50, z-score = -1.25, R) >droSec1.super_0 18468529 96 + 21120651 GCAGCAGCCAGAGUGGGCGUG--------GCUGGCCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGACCAGAAUC--------------- ((.((.((((((.((((((.(--------(....))))))))))))))..)).((((........))))))......(((...(((((....))))).)))...--------------- ( -43.00, z-score = -3.28, R) >droSim1.chr3R 17918267 96 + 27517382 GCAGCAGCCAGAGUGGGCGUG--------GCUGGCCCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGACCAGAAUC--------------- ((.((.((((((.((((((.(--------(....))))))))))))))..)).((((........))))))......(((...(((((....))))).)))...--------------- ( -43.00, z-score = -3.28, R) >consensus GCAGCAGCCAGAGUGGGCGUG________GCUGG_CCGCCCGUCUGGCGAGCGUCGCAUGCAUAAGCGAGCAUAAAAUCUUAUGUCCGUUUCCGGACCAGAAUC_______________ ............((((((((................((((.....)))).))))).)))....(((((..(((((....)))))..)))))............................ (-17.43 = -17.76 + 0.33)
| Location | 18,110,826 – 18,110,922 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Shannon entropy | 0.45333 |
| G+C content | 0.57654 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -18.62 |
| Energy contribution | -18.39 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18110826 96 - 27905053 ---------------GAUUCUGGUCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGCGACGCUCGCCAGACGGGCGGGCCAGC--------CACGCCCACUCUGGCUGCUGC ---------------...(((.(((((....)))))...)))......((((((........)))).))..((((((.(((((((....)--------).)))))..))))))...... ( -45.00, z-score = -4.29, R) >droGri2.scaffold_15074 1246371 104 - 7742996 GGUUGCUGUUGUUGCAAAAGGCAACAGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGUGACGCCCGCCAGAC-GGCGC--------------CACGCCUACUGCCGCAGCAGC ....((((((((.(((..((((....(....)(..(((((....)))))..)((....))..(((.((((........-)))).--------------)))))))..))).)))))))) ( -39.10, z-score = -1.72, R) >droMoj3.scaffold_6540 27803042 82 - 34148556 ----------------------UGCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGUGACGCUCGCCAGAC-GGUGC--------------CACGCCCACUCAUCUAACAAG ----------------------(((((....))).))..((((...((((........))))(((.((..((((....-)))).--------------..))..)))..))))...... ( -19.10, z-score = -0.30, R) >droVir3.scaffold_13047 10851089 97 + 19223366 UGUUUUUGUUGUUGC----UGCAGCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGUGACGCUCGCCAGAC-GGCGC--------------CACGCCCACGGCAACAAC--- .......((((((((----((..((((....))).)..........((((........))))(((.(((.((.....)-)))).--------------))).....))))))))))--- ( -32.40, z-score = -1.31, R) >droAna3.scaffold_13340 9095573 103 - 23697760 ---------------GAUCCUGGUCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGUGACGCUCGCCAGACGGGCGGACGGGCUCGCGUGGCACGCCC-CCUUAGACGUUGC ---------------((((...(((((....)))))....))))..((((........))))(..(((..((((.....))))..)(((...(((.....))).)-))......))..) ( -35.70, z-score = -0.26, R) >droEre2.scaffold_4820 500101 96 + 10470090 ---------------GAUUCUGGUCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGCGACGCUCGCCAGACGGGCGGCCCAGC--------CACGCCCACUUUAGCUGCUGC ---------------...(((((((((....)))))............((((((........)))).))....)))).(((((.......--------..))))).............. ( -33.50, z-score = -1.42, R) >droYak2.chr3R 18954236 96 - 28832112 ---------------GAUUCUGGUCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGCGACGCUCGCCAGACGGGCGGACCAGC--------CACGCCCACUUUUGGUGCUGC ---------------...(((.(((((....)))))...)))......((((((........)))).)).(((((((.(((((.......--------..)))))...))))))).... ( -37.90, z-score = -3.01, R) >droSec1.super_0 18468529 96 - 21120651 ---------------GAUUCUGGUCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGCGACGCUCGCCAGACGGGCGGGCCAGC--------CACGCCCACUCUGGCUGCUGC ---------------...(((.(((((....)))))...)))......((((((........)))).))..((((((.(((((((....)--------).)))))..))))))...... ( -45.00, z-score = -4.29, R) >droSim1.chr3R 17918267 96 - 27517382 ---------------GAUUCUGGUCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGCGACGCUCGCCAGACGGGCGGGCCAGC--------CACGCCCACUCUGGCUGCUGC ---------------...(((.(((((....)))))...)))......((((((........)))).))..((((((.(((((((....)--------).)))))..))))))...... ( -45.00, z-score = -4.29, R) >consensus _______________GAUUCUGGUCCGGAAACGGACAUAAGAUUUUAUGCUCGCUUAUGCAUGCGACGCUCGCCAGACGGGCGG_CCAGC________CACGCCCACUCUAGCUGCUGC ........................(((....)))..............((((((........)))).)).((((.....)))).................................... (-18.62 = -18.39 + -0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:51 2011