
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,262,086 – 8,262,201 |
| Length | 115 |
| Max. P | 0.898555 |

| Location | 8,262,086 – 8,262,201 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 64.24 |
| Shannon entropy | 0.80970 |
| G+C content | 0.42404 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -14.31 |
| Energy contribution | -13.94 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.34 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
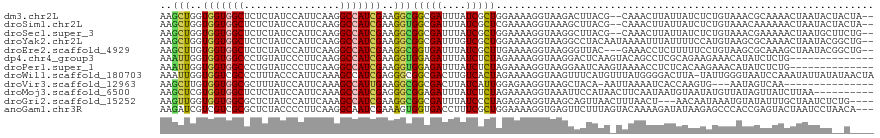
>dm3.chr2L 8262086 115 - 23011544 AAGCUGGUGGUGGCUCUCUAUCCAUUCAAGGCCAUCGAAGGCGGCGAUUUAUCGCUGGAAAAGGUAAGACUUACG--CAAACUUAUUAUCUCUGUAAACGCAAAACUAAUACUACUA-- ..(((..((((((((..............))))))))..)))(((((....)))))(((.(..(((((.......--....)))))..).)))........................-- ( -23.34, z-score = 0.26, R) >droSim1.chr2L 8046267 115 - 22036055 AAGCUGGUGGUGGCUCUCUAUCCAUUCAAGGCCAUCGAAGGUGGCGAUUUAUCGCUCGAAAAGGUAAAGCUUACG--CAAACUUAUUAUCUCUGUAAACAAAAAACUAAUACUACUA-- ..(((..((((((((..............))))))))..)))(((((....)))))......((((..((....)--)....((((.......))))............))))....-- ( -20.04, z-score = 0.55, R) >droSec1.super_3 3758728 115 - 7220098 AAGCUGGUGGUGGCUCUCUAUCCAUUCAAGGCCAUCGAAGGCGGCGAUUUAUCGCUGGAAAAGGUAAGGCUUACG--CAAACUUAUUAUCUCUGUAAACGAAAAACUAAUGCUUCUG-- ..(((..((((((((..............))))))))..)))(((((....)))))(((.(..(((((((....)--)...)))))..).)))........................-- ( -24.84, z-score = 0.46, R) >droYak2.chr2L 10897777 117 - 22324452 AAGCUGGUGGUGGCUCUCUAUCCAUUCAAGGCCAUCGAAGGCGGCGAUUUGUCGCUGGAAAAGGUAAGGCCUACAAUAAAAUUUAUUUUCCAUGUAAGCGCAAAACUAAUACGGCUG-- .(((((.((((((((..............))))))))...(((((((....))))(((((((((.....)))..............))))))......)))..........))))).-- ( -30.07, z-score = -0.22, R) >droEre2.scaffold_4929 17171070 114 - 26641161 AAGCUUGUGGUGGCUCUCUAUCCAUUCAAGGCCAUCGAAGGCGGUGAUUUAUCGCUUGAAAAGGUAAGGGUUAC---GAAACCUCUUUUUCCUGUAAGCGCAAAGCUAAUACGGCUG-- .((((.(((.(((((..........(((..(((......)))..))).....((((((...(((.(((((((..---..)))).)))...))).))))))...))))).))))))).-- ( -33.80, z-score = -1.28, R) >dp4.chr4_group3 7776445 105 + 11692001 AAAUUGGUGGUGGCCCUGUAUCCCUUCAAGGCCAUCGAAGGUGGAGAUUUAUCUCUAGAAAAGGUAAGGACUCAAGUACAGCCUCGCAGAAGAAACAUAUCUCUG-------------- ..(((..((((((((.((........)).))))))))..)))((((((...(((((.(...((((....((....))...))))..))).))).....)))))).-------------- ( -26.50, z-score = -0.11, R) >droPer1.super_1 4884586 105 + 10282868 AAAUUGGUGGUGGCCCUGUAUCCCUUCAAGGCCAUCGAAGGUGGAGAUUUAUCUCUAGAAAAGGUAAGGAAUCAAGUAAAACCUCUCACAAGAAACAUAUCUCUG-------------- ..(((..((((((((.((........)).))))))))..)))((((((.....(((.(...((((...............))))....).))).....)))))).-------------- ( -23.66, z-score = -0.12, R) >droWil1.scaffold_180703 3456921 118 + 3946847 AAAUUGGUGGUCGCCCUUUACCCAUUCAAAGCCAUCGAGGGCGGCGACUUGUCACUAGAAAAGGUAAGUUUCAUGUUUAUGGGGACUUA-UAUUGGGUAAUCCAAAUAUUAUAUAACUA .....(((.(((((((((..................))))))))).)))..............((((((..(.........)..)))))-).((((.....)))).............. ( -27.37, z-score = -0.00, R) >droVir3.scaffold_12963 16005678 100 - 20206255 AAGCUUGUGGUGGCGCUUUAUCCAUUCAAAGCCAUUGAAGGCGGCGACUUAUCAUUGGAGAAGGUAAGCUACA-AAUUAAAAUCACCAAGUG---AAUAGUCAA--------------- ..((((.(((((((................))))))).))))...((((..(((((((.((...(((......-..)))...)).)).))))---)..))))..--------------- ( -23.19, z-score = -0.51, R) >droMoj3.scaffold_6500 5449973 109 - 32352404 AAGCUCGUGGUGGCUCUCUAUCCAUUCAAAGCCAUCGAGGGCGGAGAUUUAUCUCUAGAAAAGGUAAAUUCCAUAACUUCAAUAAUGUAAUAUGUUAUAGUUAUCUUAA---------- ..((((.((((((((..............)))))))).))))((((.(((((((.......)))))))))))((((((..((((........))))..)))))).....---------- ( -28.24, z-score = -2.80, R) >droGri2.scaffold_15252 13439785 112 - 17193109 AAGUUGGUGGUGGCGCUCUAUCCAUUCAAAGCCAUCGAAGGCGGCGAUUUAUCCCUAGAGAAGGUAAGCAGUUAACUUUAACU---AACAAUAAAUGUAUAUUUGCUAAUCUCUG---- ..(((..(((((((................)))))))..)))((.((....))))((((((.((((((.(((((....)))))---.(((.....)))...))))))..))))))---- ( -23.79, z-score = -0.01, R) >anoGam1.chr3R 44330357 116 + 53272125 AAGAUCGUCGUCGCGCUCUACCCCUUCAAGGCAAUCGAAAGUGGUGACCUUUCGCUGGAAAAGGUGAGUUCUUUAGUACAAAAGAUAUAAGAGCCCACCGAGUACUAAUCCUAACA--- ..(((....)))(..(((..((((.....))....((((((.(....)))))))..))....((((.((((((...............)))))).)))))))..)...........--- ( -26.26, z-score = -0.31, R) >consensus AAGCUGGUGGUGGCUCUCUAUCCAUUCAAGGCCAUCGAAGGCGGCGAUUUAUCGCUAGAAAAGGUAAGGCUUACA_UAAAACUUAUUAUAUAUGUAAUAGCAAAACUAAUAC__C____ ..(((..(((((((................)))))))..)))(((((....)))))............................................................... (-14.31 = -13.94 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:37 2011