
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,090,677 – 18,090,781 |
| Length | 104 |
| Max. P | 0.591410 |

| Location | 18,090,677 – 18,090,781 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.30 |
| Shannon entropy | 0.18883 |
| G+C content | 0.40947 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18090677 104 - 27905053 ---UCCCUCCCUCAGAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCC------------- ---......(.((((......)))).).........((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))..------------- ( -20.90, z-score = -0.89, R) >droEre2.scaffold_4820 474638 104 + 10470090 ---UCCCUCCCUCAGAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCC------------- ---......(.((((......)))).).........((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))..------------- ( -20.90, z-score = -0.89, R) >droYak2.chr3R 18932103 104 - 28832112 ---UCCCUCCCUCAGAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCC------------- ---......(.((((......)))).).........((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))..------------- ( -20.90, z-score = -0.89, R) >droSim1.chr3R 17898150 104 - 27517382 ---UCCCUCCCUCAGAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCC------------- ---......(.((((......)))).).........((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))..------------- ( -20.90, z-score = -0.89, R) >droWil1.scaffold_181130 12230552 120 - 16660200 ACUCUUUACUACCAAAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCAGCGGGCGGGCCUA ...........((...(((((.....(((....)))((((((.((....((.(((((((((.((((....))))......))))))))).))....)).)))))))))))...))..... ( -24.90, z-score = 0.88, R) >droAna3.scaffold_13340 9077013 97 - 23697760 ----------CUCAGAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCC------------- ----------.......((..((......))..)).((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))..------------- ( -20.90, z-score = -0.82, R) >droSec1.super_0 18448630 104 - 21120651 ---UCCCUCCCUCAGAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCC------------- ---......(.((((......)))).).........((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))..------------- ( -20.90, z-score = -0.89, R) >droGri2.scaffold_15074 1224660 100 - 7742996 -------CGUUCCCGUUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCGA------------- -------......(((..((......))..))).((((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))))------------- ( -25.60, z-score = -2.13, R) >consensus ___UCCCUCCCUCAGAUGUUGUUGAUGAUGAUGAUUGUAAUCCGGUUUGUAAGUUAUGUGCUGCUUAAUUGAGUUCACAUGCACAUGACCUGAAUUCUUGAUUGCCC_____________ .................((..((......))..)).((((((.((....((.(((((((((.((((....))))......))))))))).))....)).))))))............... (-21.01 = -21.01 + -0.00)

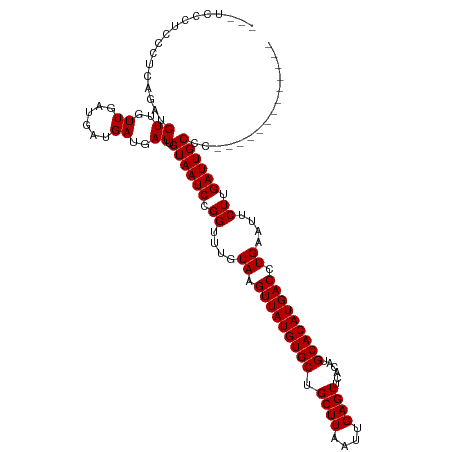
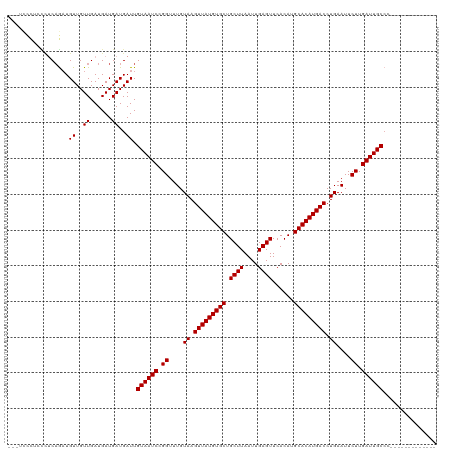
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:46 2011