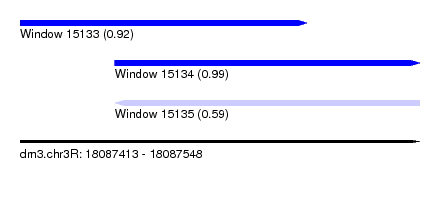
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,087,413 – 18,087,548 |
| Length | 135 |
| Max. P | 0.988294 |

| Location | 18,087,413 – 18,087,510 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.64 |
| Shannon entropy | 0.35926 |
| G+C content | 0.43367 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -14.46 |
| Energy contribution | -15.35 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
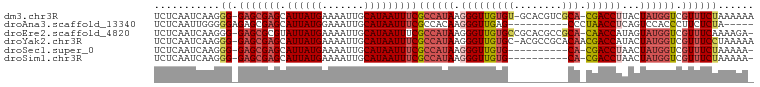
>dm3.chr3R 18087413 97 + 27905053 UCUCAAUCAAGGG-GAGCGAGCAUUAUGAAAAUUGCAUAAUUUCGCCAUAAGGGUUGUGU-GCACGUCGCA-CGACCUUACUAUGGUCGUUUCUAAAAAA ...........((-(((((((.((((((.......)))))))))((((((((((((((((-(.....))))-)))))))..)))))).))))))...... ( -30.80, z-score = -3.40, R) >droAna3.scaffold_13340 9074144 85 + 23697760 UCUCAAUUGGGGGAGAGCGAGCAUUAUGGAAAUUGCAUAAUUUCGCCACAAGGGUUGAG----------CCCUAACCUCAGUCCACCCUUCUCUA----- .......((((((((.(((((.((((((.......))))))))))).....((..((((----------.......))))..))...))))))))----- ( -26.10, z-score = -1.52, R) >droEre2.scaffold_4820 471389 97 - 10470090 UCUCAAUCAAGGG-GAGCGCGUAUUAUGAAAAUUGCAUAAUUUCGCCAUAAGGGUUGUGCCGCACGCCGCA-CAACCAUAGUAUGGUCGUUUCAAAAGA- ............(-(((((.(.((((((.......))))))..)((((((..((((((((.(....).)))-)))))....))))))))))))......- ( -29.10, z-score = -2.15, R) >droYak2.chr3R 18928768 98 + 28832112 UCUCAAUCAAGGG-GAGCGAGCAUUAUGAAAAUUGCAUAAUUUCGCCAUAAGGGUUGUGC-ACGCCGCACAACGACCAUACUAUGGUCGUUUCCUAAAAA .........(((.-.((((((.((((((.......))))))))))).......(((((((-.....)))))))((((((...)))))).)..)))..... ( -31.50, z-score = -3.47, R) >droSec1.super_0 18445431 87 + 21120651 UCUCAAUCAAGGG-GAGCGAGCAUUAUGAAAAUUGCAUAAUUUCGCCAUAAGGGUUGUG----------CA-CGACCUAACUAUGGUCGUUUCUAAAAA- ...........((-(((((((.((((((.......)))))))))((((((.((((((..----------..-))))))...)))))).)))))).....- ( -23.10, z-score = -2.20, R) >droSim1.chr3R 17894925 87 + 27517382 UCUCAAUCAAGGG-GAGCGAGCAUUAUGAAAAUUGCAUAAUUUCGCCAUAAGGGUUGUG----------CA-CGACCUAACUAUGGUCGUUUCUAAAAA- ...........((-(((((((.((((((.......)))))))))((((((.((((((..----------..-))))))...)))))).)))))).....- ( -23.10, z-score = -2.20, R) >consensus UCUCAAUCAAGGG_GAGCGAGCAUUAUGAAAAUUGCAUAAUUUCGCCAUAAGGGUUGUG__________CA_CGACCUUACUAUGGUCGUUUCUAAAAA_ ...(((((....(.(.(((((.((((((.......)))))))))))).)...)))))...............(((((.......)))))........... (-14.46 = -15.35 + 0.89)
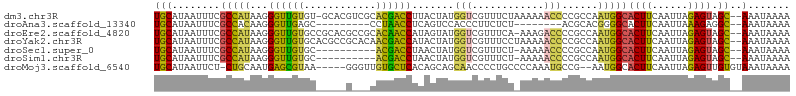
| Location | 18,087,445 – 18,087,548 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.54613 |
| G+C content | 0.43761 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -11.08 |
| Energy contribution | -10.17 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
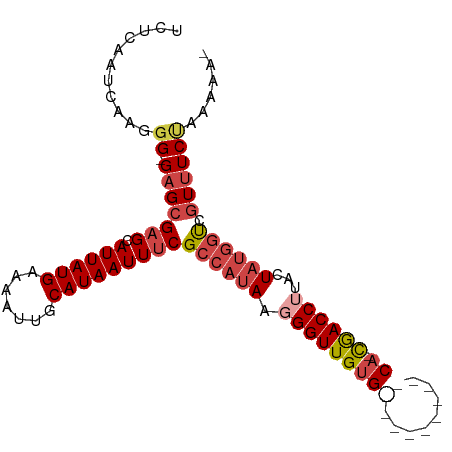
>dm3.chr3R 18087445 103 + 27905053 UGCAUAAUUUCGCCAUAAGGGUUGUGU-GCACGUCGCACGACCUUACUAUGGUCGUUUCUAAAAAACCCCGCCAAUGGCACUUCAAUUAGAGUAGC--AAAUAAAA (((........(((((.((((((((((-(.....)))))))))))....((((.((((.....))))...)))))))))((((......)))).))--)....... ( -30.00, z-score = -2.93, R) >droAna3.scaffold_13340 9074177 87 + 23697760 UGCAUAAUUUCGCCACAAGGGUUGAGC---------CCUAACCUCAGUCCACCCUUCUCU--------ACGCACGGGGCACUUCAAUUAAAGAGGC--AAAUAAAA ...........(((....((..((((.---------......))))..)).(((......--------......)))................)))--........ ( -17.80, z-score = -0.26, R) >droEre2.scaffold_4820 471421 103 - 10470090 UGCAUAAUUUCGCCAUAAGGGUUGUGCCGCACGCCGCACAACCAUAGUAUGGUCGUUUCA-AAAGACCCCGCCAAUGGCACUUCAAUUAGAGUAGC--AAAUAAAA (((........(((((...((((((((.(....).))))))))...((..((((......-...))))..))..)))))((((......)))).))--)....... ( -31.20, z-score = -3.08, R) >droYak2.chr3R 18928800 104 + 28832112 UGCAUAAUUUCGCCAUAAGGGUUGUGCACGCCGCACAACGACCAUACUAUGGUCGUUUCCUAAAAACCCCGCCAAUGGCACUUCAAUUAGAGUAGC--AAAUAAAA (((........(((((..(((((((((.....))))(((((((((...))))))))).......))))).....)))))((((......)))).))--)....... ( -32.70, z-score = -4.04, R) >droSec1.super_0 18445463 93 + 21120651 UGCAUAAUUUCGCCAUAAGGGUUGUGC----------ACGACCUAACUAUGGUCGUUUCU-AAAAACCCCGCCAAUGGCACUUCAAUUAGAGUAGC--AAAUAAAA (((........(((((..(((((....----------((((((.......))))))....-...))))).....)))))((((......)))).))--)....... ( -25.40, z-score = -2.85, R) >droSim1.chr3R 17894957 93 + 27517382 UGCAUAAUUUCGCCAUAAGGGUUGUGC----------ACGACCUAACUAUGGUCGUUUCU-AAAAACCCCGCCAAUGGCACUUCAAUUAGAGUAGC--AAAUAAAA (((........(((((..(((((....----------((((((.......))))))....-...))))).....)))))((((......)))).))--)....... ( -25.40, z-score = -2.85, R) >droMoj3.scaffold_6540 27778216 98 + 34148556 UGCAUAAUUCU-CUGCAAUGAGCGUAA-----GGGUUGUGCUCACAGCAGCAACCCCUGCCCCAAAUGCCG--AAUGGCACUUCAAUUAGAGUUGUGUAAAUAAAA (((((((((((-......((.(.(((.-----(((((((((.....))).)))))).)))).))..((((.--...))))........)))))))))))....... ( -31.60, z-score = -3.01, R) >consensus UGCAUAAUUUCGCCAUAAGGGUUGUGC_____G___CACGACCUUACUAUGGUCGUUUCU_AAAAACCCCGCCAAUGGCACUUCAAUUAGAGUAGC__AAAUAAAA .((........(((((...((((((............)))))).......(((............)))......)))))((((......)))).)).......... (-11.08 = -10.17 + -0.91)
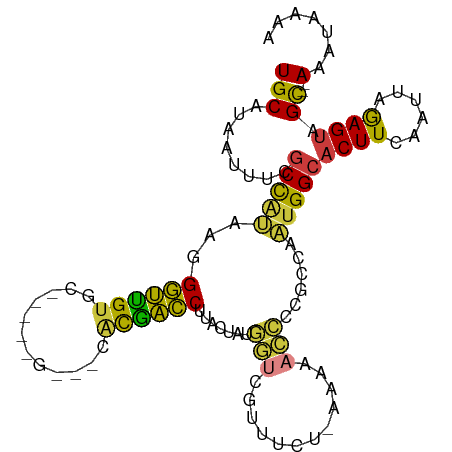
| Location | 18,087,445 – 18,087,548 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.54613 |
| G+C content | 0.43761 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -11.98 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
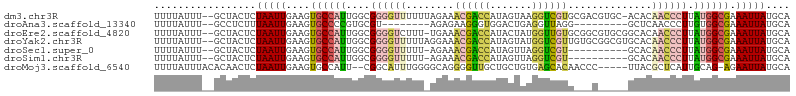
>dm3.chr3R 18087445 103 - 27905053 UUUUAUUU--GCUACUCUAAUUGAAGUGCCAUUGGCGGGGUUUUUUAGAAACGACCAUAGUAAGGUCGUGCGACGUGC-ACACAACCCUUAUGGCGAAAUUAUGCA .......(--((.....(((((....((((((....((((((..........((((.......))))((((.....))-))..)))))).)))))).))))).))) ( -28.00, z-score = -1.27, R) >droAna3.scaffold_13340 9074177 87 - 23697760 UUUUAUUU--GCCUCUUUAAUUGAAGUGCCCCGUGCGU--------AGAGAAGGGUGGACUGAGGUUAGG---------GCUCAACCCUUGUGGCGAAAUUAUGCA .....(((--(((.(..(((((..(((.((((...(..--------...)..))).).)))..)))))((---------(.....)))..).))))))........ ( -23.10, z-score = -0.14, R) >droEre2.scaffold_4820 471421 103 + 10470090 UUUUAUUU--GCUACUCUAAUUGAAGUGCCAUUGGCGGGGUCUUU-UGAAACGACCAUACUAUGGUUGUGCGGCGUGCGGCACAACCCUUAUGGCGAAAUUAUGCA .......(--((.....(((((....((((((.((...((((...-......)))).......((((((((.(....).)))))))))).)))))).))))).))) ( -30.50, z-score = -1.27, R) >droYak2.chr3R 18928800 104 - 28832112 UUUUAUUU--GCUACUCUAAUUGAAGUGCCAUUGGCGGGGUUUUUAGGAAACGACCAUAGUAUGGUCGUUGUGCGGCGUGCACAACCCUUAUGGCGAAAUUAUGCA .......(--((.....(((((....((((((....((((((.......(((((((((...)))))))))((((.....)))))))))).)))))).))))).))) ( -33.10, z-score = -1.90, R) >droSec1.super_0 18445463 93 - 21120651 UUUUAUUU--GCUACUCUAAUUGAAGUGCCAUUGGCGGGGUUUUU-AGAAACGACCAUAGUUAGGUCGU----------GCACAACCCUUAUGGCGAAAUUAUGCA .......(--((.....(((((....((((((....((((((...-....((((((.......))))))----------....)))))).)))))).))))).))) ( -25.70, z-score = -1.77, R) >droSim1.chr3R 17894957 93 - 27517382 UUUUAUUU--GCUACUCUAAUUGAAGUGCCAUUGGCGGGGUUUUU-AGAAACGACCAUAGUUAGGUCGU----------GCACAACCCUUAUGGCGAAAUUAUGCA .......(--((.....(((((....((((((....((((((...-....((((((.......))))))----------....)))))).)))))).))))).))) ( -25.70, z-score = -1.77, R) >droMoj3.scaffold_6540 27778216 98 - 34148556 UUUUAUUUACACAACUCUAAUUGAAGUGCCAUU--CGGCAUUUGGGGCAGGGGUUGCUGCUGUGAGCACAACCC-----UUACGCUCAUUGCAG-AGAAUUAUGCA ..............((((.....(((((((...--.))))))).((((((((((((.(((.....)))))))))-----))..)))).....))-))......... ( -32.20, z-score = -2.47, R) >consensus UUUUAUUU__GCUACUCUAAUUGAAGUGCCAUUGGCGGGGUUUUU_AGAAACGACCAUAGUAAGGUCGUG___C_____GCACAACCCUUAUGGCGAAAUUAUGCA .................(((((....((((((.((.((.((.........((((((.......))))))..........))....)))).)))))).))))).... (-11.98 = -12.38 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:46 2011