| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,081,248 – 18,081,351 |
| Length | 103 |
| Max. P | 0.782032 |

| Location | 18,081,248 – 18,081,351 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.49411 |
| G+C content | 0.47373 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -18.81 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18081248 103 - 27905053 -------CCUUUGCUAUACGUUCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACA-GCAGUCGGGUGAAGCAAGCGAGCAGGA--- -------((((((((.(((((.(((((((((((........)))).))))))).)).(((((((.....)))))))(((....-...)))..))).)))))......))).--- ( -29.20, z-score = -0.51, R) >droAna3.scaffold_13340 9068259 90 - 23697760 UUUCUGCUGUGUCCUGUGCGCUCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAA----CAGUCGGGA-------------------- .((((((((((((......(..(((((((((((........)))).)))))))..).(((((((.....)))))))))).)----))).)))))-------------------- ( -29.30, z-score = -1.62, R) >droEre2.scaffold_4820 465188 86 + 10470090 -------CCUUUGCUAUACGUUCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACA-GCAGUCGGGA-------------------- -------((((((((....((.(((((((((((........)))).))))))).)).(((((((.....)))))))......)-))))..))).-------------------- ( -26.10, z-score = -1.98, R) >droYak2.chr3R 18922362 110 - 28832112 -CCUUUGCCUUUGCUAUACGUUCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACA-GCAGUCGGGAAAAGCAUGCGAACAGGAC-- -(((((((...((((.....(((((((.((((((((((((.....)).)))))))).(((((((.....))))))).......-.)).))))))).)))).))))..)))..-- ( -33.20, z-score = -1.64, R) >droSec1.super_0 18439789 103 - 21120651 -------CCUUUGCUAUACGUUCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACA-GCAGUCGGGUGAAGCAAGCGAGCAGGA--- -------((((((((.(((((.(((((((((((........)))).))))))).)).(((((((.....)))))))(((....-...)))..))).)))))......))).--- ( -29.20, z-score = -0.51, R) >droSim1.chr3R 17888048 103 - 27517382 -------CCUUUGCUAUACGUUCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACA-GCAGUCGGGCGAAGCAAGCGAGCAGGA--- -------((((((((....((.(((((((((((........)))).))))))).)).(((((((.....)))))))(((....-...))).))))))((......)).)).--- ( -29.10, z-score = -0.34, R) >droWil1.scaffold_181130 12220996 82 - 16660200 ---------------CCAC-UGCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACAGGCAACCCCGCAGGA---------------- ---------------((..-(((((((((((((........)))).))))))))).....(((((...((((............))))..))))))).---------------- ( -24.20, z-score = -1.19, R) >droVir3.scaffold_13047 10822116 102 + 19223366 -----------GGUGCUACGUCCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACAGGCAGUGCCCAACCACAGC-GCAGCACAGGA -----------.(((((.(((((((((((((((........)))).)))))))....(((((((.....)))))))))).....((.(((......))).))-).))))).... ( -31.50, z-score = -0.51, R) >droGri2.scaffold_15074 1215905 103 - 7742996 -----------GCUGCUACGUCCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACAGGCAGUUCCCAACCACAGCAGCAGCACAGGA -----------((((((.((....)).(((((............(((((((.((...(((((((.....))))))).))..)))))))......)))))))))))......... ( -32.87, z-score = -1.54, R) >consensus _______CCUUUGCUAUACGUUCUCGAGUGGUAUUUAACAUUAUCUGCUUGAGUACAUGUUGCGGUAUUUGCAGCAGACAACA_GCAGUCGGGAAAAGCA_G_GA_CAGGA___ ...................((.(((((((((((........)))).))))))).)).(((((((.....)))))))(((........)))........................ (-18.81 = -19.28 + 0.47)

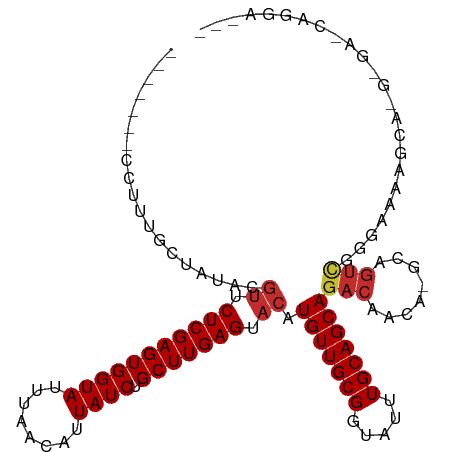

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:43 2011