| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,249,590 – 8,249,647 |
| Length | 57 |
| Max. P | 0.762155 |

| Location | 8,249,590 – 8,249,647 |
|---|---|
| Length | 57 |
| Sequences | 10 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 70.22 |
| Shannon entropy | 0.60776 |
| G+C content | 0.56537 |
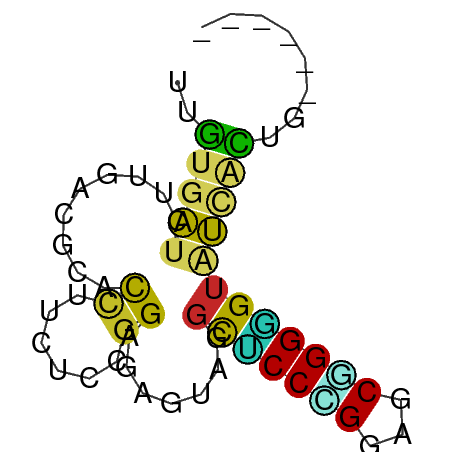
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -8.21 |
| Energy contribution | -7.27 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 2.07 |
| Mean z-score | -1.59 |
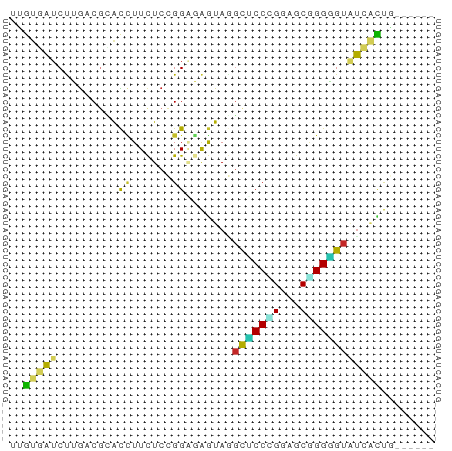
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.762155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
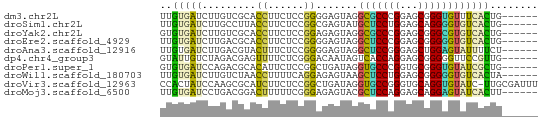
>dm3.chr2L 8249590 57 + 23011544 UUGUGAUCUUGUCGCACCUUCUCCGGGGAGUAGGCGCCCGGAGCGGGUGUUUCACUG------ ..((((.......((((((.(((((((..(....).))))))).)))))).))))..------ ( -25.60, z-score = -2.59, R) >droSim1.chr2L 8032218 57 + 22036055 UUGUGAUCUUGCCUUACCUUCUCCGGCGAGUAUGCUCCUGGAGCAGGGGUGUCACUG------ ..(((((...(((...(((.((((((.(((....))))))))).)))))))))))..------ ( -23.90, z-score = -3.04, R) >droYak2.chr2L 10885053 57 + 22324452 GUGUGAUCUUGUCGCACCUUCUCCGGAGAGUAGGCGCCCGGAGCGGGCGUGUCACUG------ (((((((...)))))))(((.....)))((((.(((((((...))))))).).))).------ ( -21.90, z-score = -0.86, R) >droEre2.scaffold_4929 17158317 57 + 26641161 UUGUGAUCUUGACGCACCUUCUCCGGGGAGUAGGCUCCCGGAGCGGGGGUGUCACUG------ .........((((((.(((.(((((((.((....))))))))).))).))))))...------ ( -28.80, z-score = -3.00, R) >droAna3.scaffold_12916 10164556 57 - 16180835 UUGUGAUCUUGACGUACUUUCUCCGGGGAGUAGGCUCCGGGAGCUGGAGUAUUUUCU------ .............(((((((((((..((((....)))).))))..))))))).....------ ( -15.40, z-score = 0.12, R) >dp4.chr4_group3 7764791 57 - 11692001 GUAUUGUCUAGACGAGUUUUCUCGGGACAAUAGUCACCAGGAGCGGGGGUUCCGUUG------ .((((((((...((((....)))))))))))).......(((((....)))))....------ ( -18.40, z-score = -1.88, R) >droPer1.super_1 4873227 57 - 10282868 GUGUGAUCCAGACGCACAUUCUCCGGCUGAUAGGUGCCCGGUGCGGGUGUAUCGCUG------ ..(((((.((..(((((.......(((........)))..)))))..)).)))))..------ ( -19.60, z-score = -0.45, R) >droWil1.scaffold_180703 3440388 57 - 3946847 UUGUGAUCUUGUCUAACCUUUUCAGGAGAGUAAGCUCCUGGAGCGGGGGUGUCACUA------ ..(((((....(((..((.(((((((((......))))))))).))))).)))))..------ ( -18.40, z-score = -1.20, R) >droVir3.scaffold_12963 15986570 62 + 20206255 CCACUAUCCAAGCGCAUCUUCUCCGGCUGAUAGGUGCCGGGUGCAGGUGUAUC-UUGCGAUUU .....(((((((.((((((.(((((((........)))))).).))))))..)-))).))).. ( -22.00, z-score = -1.79, R) >droMoj3.scaffold_6500 5432900 57 + 32352404 UUGUGAUCCUGACGGACUUUUUCGGGAGAGUACGCUCCAGGAGCAGGAGUAUCACUU------ ..(((((((((.((((....))))((((......)))).....)))))...))))..------ ( -17.30, z-score = -1.17, R) >consensus UUGUGAUCUUGACGCACCUUCUCCGGAGAGUAGGCUCCCGGAGCGGGGGUAUCACUG______ ..(((((.........((......)).......(((((((...))))))))))))........ ( -8.21 = -7.27 + -0.94)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:25:36 2011