
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,057,214 – 18,057,313 |
| Length | 99 |
| Max. P | 0.930269 |

| Location | 18,057,214 – 18,057,313 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 58.62 |
| Shannon entropy | 0.80373 |
| G+C content | 0.35089 |
| Mean single sequence MFE | -25.09 |
| Consensus MFE | -5.33 |
| Energy contribution | -5.56 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
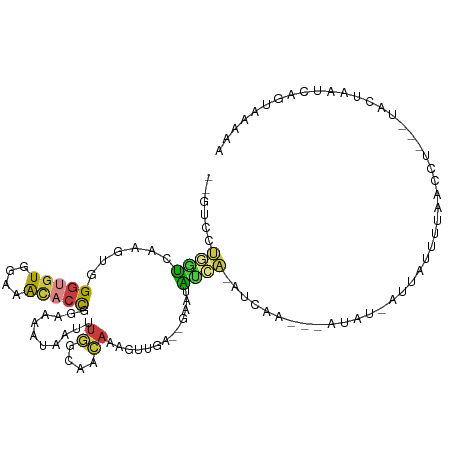
>dm3.chr3R 18057214 99 - 27905053 --GUCCUGGUCAAGUGGGUGUGGAAACACCGGAAAAUAAUUUGGCAACAAAGUUGA--GGUUAUCA-AUCAA---AUAU-AUUAUUUUAACCU----ACUAAUCAGUAAAUA --...(((((..((((((((((....)))...(((((((((((....))))(((((--.....)))-))...---....-.))))))).))))----))).)))))...... ( -26.50, z-score = -3.21, R) >droAna3.scaffold_13340 9054888 86 - 23697760 --AACCAGGCUAAGAGGGUAUUCAUGCACUGAGAAAAGUUAAGCUGAGGAUUAUGA--AAAUGCCACAUUAUUAUACAGAUCAAUCUCCA---------------------- --.....((.(((...((((((((((..((.((..........)).))...)))))--..)))))...)))......(((....))))).---------------------- ( -10.30, z-score = 1.72, R) >droEre2.scaffold_4820 451404 104 + 10470090 --GUCCUGGUCAAGUGGGUGUGGAAACACCGGAAAAUAAUUUGGCAACAAAGCUGGUUGAAUGCGA-AAUA----AUAU-GUUCUUUUCGCCUACCUACUAAUCAGGAAAAU --.(((((((..((((((((.(....)(((((.......((((....)))).))))).....((((-((..----....-.....)))))).)))))))).))))))).... ( -33.90, z-score = -3.97, R) >droYak2.chr3R 18908469 109 - 28832112 --GUCCUGGUCAAGUGGGCGUGGAAACACCGGAAAAUAAUUUGGUAACAAAGUGGUUUGCAUAGGA-ACUAAACUAUAUAGUUCCUUUAACCUAUUUGCUAAUCAGGAAAAC --.(((((((((((((((.(((....))).(.(((....((((....))))....))).)..((((-((((.......))))))))....))))))))...))))))).... ( -34.60, z-score = -3.96, R) >droSec1.super_0 18426243 102 - 21120651 --GUCCUGGUCAAGUGGGUGUGGAAACACCGGAAAAUAAUGUGGCAACAAAGUUGA--GGAAAUCA-AUCAA---AUAU-AUUAUUUUAACCU-UCUACUAAUCAGUAAAAA --...(((((..(((((((((....)))))(((((((((((((....)...(((((--.....)))-))...---..))-))))))))..)).-..)))).)))))...... ( -24.30, z-score = -2.35, R) >droSim1.chr3R 17874108 99 - 27517382 --GUCCUGGUCAAGUGGGUGUGGAAACACCGGAAAAUAAUAUGGCAACAAAGUUGA--GAAAAUCA-AUCAA---AUAU-AUUAUUUUAACCU----ACUAAUCAGUAAAAA --...(((((..((((((((((....)))...(((((((((((....)...(((((--.....)))-))...---..))-)))))))).))))----))).)))))...... ( -27.20, z-score = -4.32, R) >droWil1.scaffold_181114 18188 105 + 73031 AUGACUCAACUGAGAAGAUGUGGCAAUCGCGACAUGUAAUUUUAUUGGAAAAAUUA--GUAAGUUGUUUGAUUCUGUAA-AUAGCAAUAAUUUAUUUGACAAGAACUA---- ....(((....))).(.((((.((....)).)))).)..((((.(..((.((((((--....((((((((......)))-)))))..)))))).))..).))))....---- ( -18.80, z-score = -0.47, R) >consensus __GUCCUGGUCAAGUGGGUGUGGAAACACCGGAAAAUAAUUUGGCAACAAAGUUGA__GAAUAUCA_AUCAA___AUAU_AUUAUUUUAACCU___UACUAAUCAGUAAAAA ................(((((....))))).................................................................................. ( -5.33 = -5.56 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:38 2011