| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,047,104 – 18,047,223 |
| Length | 119 |
| Max. P | 0.943858 |

| Location | 18,047,104 – 18,047,223 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Shannon entropy | 0.39621 |
| G+C content | 0.50169 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -22.54 |
| Energy contribution | -25.83 |
| Covariance contribution | 3.29 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
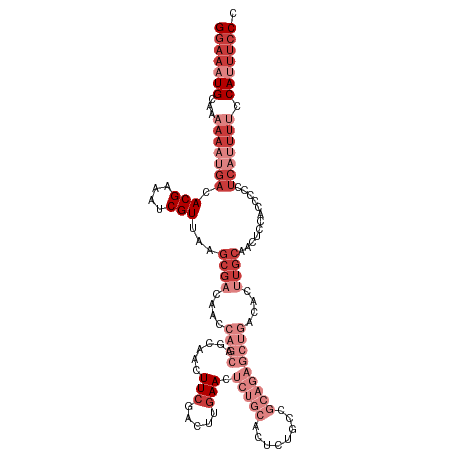
>dm3.chr3R 18047104 119 + 27905053 GGGAAAUGGAAAAUGAGGGGGAGGAGUUGCAAGUGUCAGCUCUGCGGCAGAGUGCAGAGUUCAAGUCGAAGUUGCUGCUGGUUGUCGCUUAACGAUUUCGUGUCAUUUUUUGCAUUUCC .(((((((((((((((.(.(((((((((((....).)))))))(((((((...((((..(((.....)))....))))...)))))))........))).).))))))))..))))))) ( -42.20, z-score = -2.78, R) >droSim1.chr3R 17863309 119 + 27517382 GGGAAAUGGAAAAUGAGGGGGAGGAGUUGCAAGUGUCAGCUCUGCGGCAGAGUGCAGAGUUCAAGUCGAAGUUGCUGCUGGUUGUCGCUUAACGAUUUCGUGUCAUUUUUUGCAUUUCC .(((((((((((((((.(.(((((((((((....).)))))))(((((((...((((..(((.....)))....))))...)))))))........))).).))))))))..))))))) ( -42.20, z-score = -2.78, R) >droSec1.super_0 18416358 119 + 21120651 GGGAAAUGGAAAAUGAGGGGGAGGAGUUGCAAGUGUCAGCUCUGCGGCAGAGUGCAGAGUUCAAGUCGAAGUUGCUGCUGGUUGUCGCUUAACGAUUUCGUGUCAUUUUUUGCAUUUCC .(((((((((((((((.(.(((((((((((....).)))))))(((((((...((((..(((.....)))....))))...)))))))........))).).))))))))..))))))) ( -42.20, z-score = -2.78, R) >droYak2.chr3R 18898307 119 + 28832112 GGGAAAUGGAAAAUGAGGGGGUGGUGCUGCAAGUGUCAGCUCUGCGGCAGAGUGCAGAGUUCAAGUCGAAGUUGCUGCUGGUUGUCGCUUAACGAUUUCGUGUCAUUUUUUGCAUUUCC .(((((((((((((((((((.(((..(.....)..))).))))(((((((...((((..(((.....)))....))))...)))))))...(((....))).))))))))..))))))) ( -41.50, z-score = -2.19, R) >droEre2.scaffold_4820 441208 119 - 10470090 GGGAAAUGGAAAAUGAGGGGGUGGAGUUGCAAGUGUCAGCUCUGCGGCAGAGUGCAGAGUUCAAGUCGAAGUUGCUGCUGGUUGUCGCUUAACGAUUUCGUGUCAUUUUUUGCAUUUCC .(((((((((((((((.(.((.((((((((....).)))))))(((((((...((((..(((.....)))....))))...))))))).........)).).))))))))..))))))) ( -40.60, z-score = -2.36, R) >dp4.chr2 12237807 98 + 30794189 GGGGGGUGUGGGGGCAGGGAGGACUGUUGCAAGUGUCAG-------GCGGAG-GCAGAGUUCAAGUCGAAGU-------------CGCUUAACGAUUCCGUGUCAUUUUUUGCAUUUCC .((..(((..((((((.((((.(((......)))((.((-------((((.(-((.........)))....)-------------))))).))..)))).))))....))..)))..)) ( -30.40, z-score = -1.11, R) >droMoj3.scaffold_6540 27745019 97 + 34148556 UGCAACUGGCAACAGGGGCCGUGGCAUUUCCGUUGCCUG------GGCAC-GCGCUU-GUUAGAAGCGAAGCU--------------CUUAACGAUAUCGUGUCAUUUUUUGCAUUUCC ((((((.((((((.(((((....))...))))))))).)------(((((-(...((-((((..(((...)))--------------..))))))...)))))).....)))))..... ( -33.60, z-score = -1.62, R) >consensus GGGAAAUGGAAAAUGAGGGGGAGGAGUUGCAAGUGUCAGCUCUGCGGCAGAGUGCAGAGUUCAAGUCGAAGUUGCUGCUGGUUGUCGCUUAACGAUUUCGUGUCAUUUUUUGCAUUUCC .(((((((((((((((.(.((((..((((..((((.(((((..(((((((..((............))...))))))).))))).))))))))..)))).).))))))))..))))))) (-22.54 = -25.83 + 3.29)


| Location | 18,047,104 – 18,047,223 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Shannon entropy | 0.39621 |
| G+C content | 0.50169 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -9.89 |
| Energy contribution | -15.14 |
| Covariance contribution | 5.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18047104 119 - 27905053 GGAAAUGCAAAAAAUGACACGAAAUCGUUAAGCGACAACCAGCAGCAACUUCGACUUGAACUCUGCACUCUGCCGCAGAGCUGACACUUGCAACUCCUCCCCCUCAUUUUCCAUUUCCC (((((((...(((((((.(((....)))...((((....((((......(((.....))).(((((........)))))))))....))))............))))))).))))))). ( -28.00, z-score = -3.41, R) >droSim1.chr3R 17863309 119 - 27517382 GGAAAUGCAAAAAAUGACACGAAAUCGUUAAGCGACAACCAGCAGCAACUUCGACUUGAACUCUGCACUCUGCCGCAGAGCUGACACUUGCAACUCCUCCCCCUCAUUUUCCAUUUCCC (((((((...(((((((.(((....)))...((((....((((......(((.....))).(((((........)))))))))....))))............))))))).))))))). ( -28.00, z-score = -3.41, R) >droSec1.super_0 18416358 119 - 21120651 GGAAAUGCAAAAAAUGACACGAAAUCGUUAAGCGACAACCAGCAGCAACUUCGACUUGAACUCUGCACUCUGCCGCAGAGCUGACACUUGCAACUCCUCCCCCUCAUUUUCCAUUUCCC (((((((...(((((((.(((....)))...((((....((((......(((.....))).(((((........)))))))))....))))............))))))).))))))). ( -28.00, z-score = -3.41, R) >droYak2.chr3R 18898307 119 - 28832112 GGAAAUGCAAAAAAUGACACGAAAUCGUUAAGCGACAACCAGCAGCAACUUCGACUUGAACUCUGCACUCUGCCGCAGAGCUGACACUUGCAGCACCACCCCCUCAUUUUCCAUUUCCC (((((((...(((((((.(((....)))...((..(((....((((...(((.....))).(((((........)))))))))....)))..)).........))))))).))))))). ( -29.80, z-score = -3.57, R) >droEre2.scaffold_4820 441208 119 + 10470090 GGAAAUGCAAAAAAUGACACGAAAUCGUUAAGCGACAACCAGCAGCAACUUCGACUUGAACUCUGCACUCUGCCGCAGAGCUGACACUUGCAACUCCACCCCCUCAUUUUCCAUUUCCC (((((((...(((((((.(((....)))...((((....((((......(((.....))).(((((........)))))))))....))))............))))))).))))))). ( -28.00, z-score = -3.56, R) >dp4.chr2 12237807 98 - 30794189 GGAAAUGCAAAAAAUGACACGGAAUCGUUAAGCG-------------ACUUCGACUUGAACUCUGC-CUCCGC-------CUGACACUUGCAACAGUCCUCCCUGCCCCCACACCCCCC (((..(((((.........((((..(((((((((-------------....)).)))))....)).-.)))).-------.......)))))....))).................... ( -14.73, z-score = -0.31, R) >droMoj3.scaffold_6540 27745019 97 - 34148556 GGAAAUGCAAAAAAUGACACGAUAUCGUUAAG--------------AGCUUCGCUUCUAAC-AAGCGC-GUGCC------CAGGCAACGGAAAUGCCACGGCCCCUGUUGCCAGUUGCA .....(((((....((.((((.....((((..--------------(((...)))..))))-.....)-)))..------))((((((((....((....))..))))))))..))))) ( -29.90, z-score = -1.31, R) >consensus GGAAAUGCAAAAAAUGACACGAAAUCGUUAAGCGACAACCAGCAGCAACUUCGACUUGAACUCUGCACUCUGCCGCAGAGCUGACACUUGCAACUCCACCCCCUCAUUUUCCAUUUCCC (((((((...(((((((.(((....)))...((((....((((......(((.....))).(((((........)))))))))....))))............))))))).))))))). ( -9.89 = -15.14 + 5.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:35 2011