
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,028,278 – 18,028,372 |
| Length | 94 |
| Max. P | 0.784814 |

| Location | 18,028,278 – 18,028,372 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Shannon entropy | 0.46628 |
| G+C content | 0.38862 |
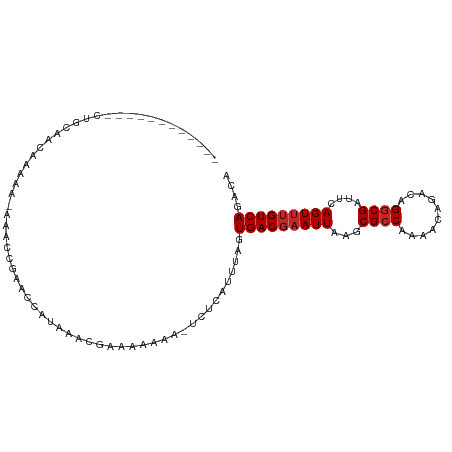
| Mean single sequence MFE | -14.74 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
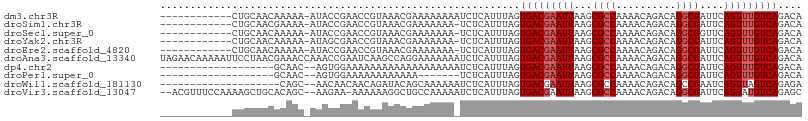
>dm3.chr3R 18028278 94 + 27905053 ------------CUGCAACAAAAA-AUACCGAACCGUAAACGAAAAAAAAUCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA ------------............-.........((....))..................(((((((((...((((..........))))....))))))))).... ( -14.70, z-score = -0.72, R) >droSim1.chr3R 17844632 93 + 27517382 ------------CUGCAACGAAAA-AUACCGAACCGUAAACGAAAAAAA-UCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA ------------............-.........((....)).......-..........(((((((((...((((..........))))....))))))))).... ( -14.70, z-score = -0.37, R) >droSec1.super_0 18397729 93 + 21120651 ------------CUGCAACAAAAA-AUACCGAACCGUAAACGAAAAAAA-UCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGGUUCAGUUUGUCAGACA ------------............-.........((....)).......-..........(((((((((((.((((..........)))).)).))))))))).... ( -16.40, z-score = -0.77, R) >droYak2.chr3R 18879000 93 + 28832112 ------------CUGCAACAAAAA-AUAGCGAACCGUAAACGAAAAAAA-UCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA ------------.(((........-...)))...((....)).......-..........(((((((((...((((..........))))....))))))))).... ( -14.80, z-score = -0.38, R) >droEre2.scaffold_4820 421156 93 - 10470090 ------------CUGCAACAAAAA-AUACCGAACCGUAAACGAAAAAAA-UCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA ------------............-.........((....)).......-..........(((((((((...((((..........))))....))))))))).... ( -14.70, z-score = -0.74, R) >droAna3.scaffold_13340 9025968 107 + 23697760 UAGAACAAAAAUUCCUAACGAAACCAAACCGAAUCAAGCCAGGAAAAAAAUCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA ...........(((((..(..................)..)))))...............(((((((((...((((..........))))....))))))))).... ( -17.07, z-score = -1.30, R) >dp4.chr2 12213720 86 + 30794189 -------------------GCAAC--AGUGGAAAAAAAAAAAAAAAAAAAUCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA -------------------.....--..................................(((((((((...((((..........))))....))))))))).... ( -14.60, z-score = -1.41, R) >droPer1.super_0 3538192 79 - 11822988 -------------------GCAAC--AGUGGAAAAAAAAAAAA-------UCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA -------------------.....--.................-------..........(((((((((...((((..........))))....))))))))).... ( -14.60, z-score = -1.22, R) >droWil1.scaffold_181130 12173727 85 + 16660200 --------------------CAGC--AACAACAACAGAUACAGCAAAAAAUCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGCCGAAUCAGUUAGUCAGAGA --------------------....--........................((((.(((.(((.(.......)))).)))...((((((.......))).))).)))) ( -8.10, z-score = 0.17, R) >droVir3.scaffold_13047 10776260 102 - 19223366 --ACGUUUCCAAAAGCUGCACAGC--AAGAA-AAAAAAGGCUGCCAAAAAUCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUAUGUCAGAGC --..((((......(((....)))--.....-......(((.((...((.((((((...)))).)).))..))))).)))).((((.((......)).))))..... ( -17.70, z-score = 0.41, R) >consensus ____________CUGCAACAAAAA_AAACCGAACCAUAAACGAAAAAAA_UCUCAUUUAGUGACGAAUUAAGCGCCAAAACAGACAGGCGAUUCAGUUUGUCAGACA ............................................................(((((((((...((((..........))))....))))))))).... (-13.03 = -13.33 + 0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:32 2011