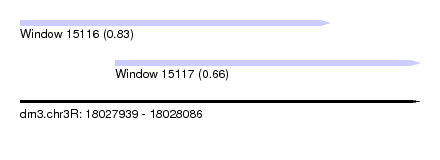
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,027,939 – 18,028,086 |
| Length | 147 |
| Max. P | 0.826771 |

| Location | 18,027,939 – 18,028,053 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Shannon entropy | 0.32404 |
| G+C content | 0.46682 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -23.82 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
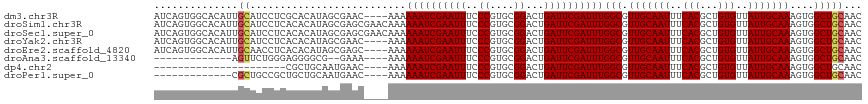
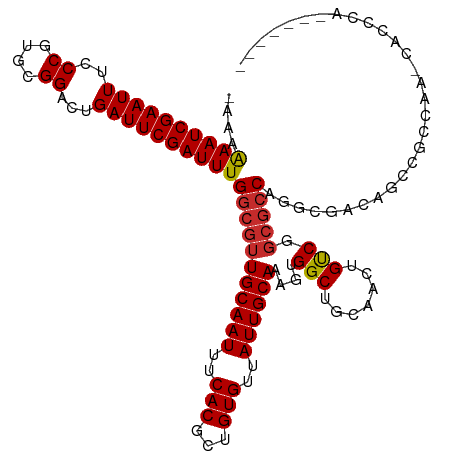
>dm3.chr3R 18027939 114 + 27905053 AUCAGUGGCACAUUGCAUCCUCGCACAUAGCGAAC----AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC ....(..((.((((((....((((.....))))..----...((((((((((..((....))...)))))))))).)).(((((((..(((...)))..)))))))))))))..)... ( -36.50, z-score = -1.91, R) >droSim1.chr3R 17844276 118 + 27517382 AUCAGUGGCACAUUGCAUCCUCACACAUAGCGAGCGAACAAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC ....(..((((.(((((.......((((((((.((((.....((((((((((..((....))...))))))))))....)))).......))))))))...))))).)).))..)... ( -36.50, z-score = -1.57, R) >droSec1.super_0 18397373 118 + 21120651 AUCAGUGGCACAUUGCAUCCUCACACAUAGCGAGCGAACAAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC ....(..((((.(((((.......((((((((.((((.....((((((((((..((....))...))))))))))....)))).......))))))))...))))).)).))..)... ( -36.50, z-score = -1.57, R) >droYak2.chr3R 18878608 114 + 28832112 AUCAGUGGCACAUUGCAUCCUCACACAUAGCGAAC----AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC ....(..((((.(((((.......((((((((...----...((((((((((..((....))...)))))))))).((...)).......))))))))...))))).)).))..)... ( -34.70, z-score = -1.72, R) >droEre2.scaffold_4820 420788 114 - 10470090 AUCAGUGGCACAUUGCAACCUCACACAUAGCGAGC----AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC ....(..((((.((((((......((((((((.((----((.((((((((((..((....))...))))))))))....)))).......))))))))..)))))).)).))..)... ( -36.80, z-score = -2.03, R) >droAna3.scaffold_13340 9025693 99 + 23697760 -------------AGUUCUGGGAGGGGCG--GAAA----AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC -------------..........(.(((.--....----...((((((((((..((....))...)))))))))).((.(((((((..(((...)))..))))))).)).))).)... ( -27.60, z-score = -0.49, R) >dp4.chr2 12213458 92 + 30794189 ----------------------CGCUGCAAUGAAC----AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC ----------------------.((.((.......----...((((((((((..((....))...)))))))))).((.(((((((..(((...)))..))))))).)).)).))... ( -27.60, z-score = -1.49, R) >droPer1.super_0 3537897 101 - 11822988 -------------CGCUGCCGCUGCUGCAAUGAAC----AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC -------------.((.((((((..((((((((((----...((((((((((..((....))...))))))))))(((((.........))))))).)))))))).)))))).))... ( -39.40, z-score = -3.93, R) >consensus AUCAGUGGCACAUUGCAUCCUCACACAUAGCGAAC____AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAAC ..............((..........................((((((((((..((....))...))))))))))(((.(((((((..(((...)))..)))))))....)))))... (-23.82 = -23.97 + 0.16)
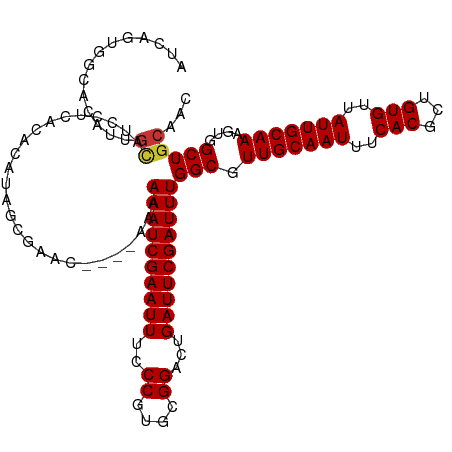
| Location | 18,027,974 – 18,028,086 |
|---|---|
| Length | 112 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.54 |
| Shannon entropy | 0.30184 |
| G+C content | 0.50948 |
| Mean single sequence MFE | -36.21 |
| Consensus MFE | -28.77 |
| Energy contribution | -29.13 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18027974 112 + 27905053 -AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCGAUAUCCACCAAACACCCA------- -...((((((((((..((....))...))))))))))(((.(((((((..(((...)))..)))))))....(((((....).))))))).((.(.....).)).........------- ( -32.30, z-score = -0.61, R) >droSim1.chr3R 17844315 112 + 27517382 -AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGAGAUAUCCACCAAACACCCA------- -...((((((((((..((....))...))))))))))(((.(((((((..(((...)))..)))))))....(((((....).))))))).(((....)))............------- ( -33.90, z-score = -1.39, R) >droSec1.super_0 18397412 112 + 21120651 -AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCGAUAUCCACCAAACACCCA------- -...((((((((((..((....))...))))))))))(((.(((((((..(((...)))..)))))))....(((((....).))))))).((.(.....).)).........------- ( -32.30, z-score = -0.61, R) >droEre2.scaffold_4820 420823 112 - 10470090 -AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCGAUAUUCGCCCAGCACCCA------- -...((((((((((..((....))...))))))))))(((((((((...((((..(((......)))..)))).)))))).)))((.((..((((.....))))..)).))..------- ( -39.70, z-score = -2.03, R) >droAna3.scaffold_13340 9025713 103 + 23697760 -AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCAGCAGCUCCU---------------- -...((((((((((..((....))...))))))))))....(((((((..(((...)))..)))))))((..(((((...((((.......)))))))))..))---------------- ( -33.80, z-score = -0.90, R) >dp4.chr2 12213471 112 + 30794189 -AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCGCAUGCCGCCUGCCGCAUG------- -...((((((((((..((....))...))))))))))(((((((((...((((..(((......)))..)))).)))))).))).(((.((((((.....)))))).)))...------- ( -42.50, z-score = -1.71, R) >droPer1.super_0 3537919 119 - 11822988 -AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCGCCAGGCGCCUGCCGCCUGCCGCCUG -...((((((((((..((....))...))))))))))(((((((((...((((..(((......)))..)))).)))))).)))((((.((((((.(((....))).)))))).)))).. ( -51.60, z-score = -3.11, R) >droWil1.scaffold_181130 12173441 99 + 16660200 AGAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCAACGG--------------------- ....((((((((((..((....))...))))))))))(((.(((((((..(((...)))..)))))))....(((((....).))))))).(....)..--------------------- ( -32.00, z-score = -0.87, R) >droGri2.scaffold_15074 1169149 101 + 7742996 -GAAGAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGCCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGCC----CCAGAACUCGAGAGCGAA-------------- -...((((((((((..((....))...))))))))))((.((((((...((((..(((......)))..)))).)))))).)).----.......(((....))).-------------- ( -27.80, z-score = -0.71, R) >consensus _AAAAAAUCGAAUUUCCCGUGCGGACUGAUUCGAUUUGGCGUUGCAAUUUCACGCUGUGUUAUUGCAAAGUGGCUGCAACUGUCGGCGCCAGGCGACAGCCGCCAA_CACCCA_______ ....((((((((((..((....))...))))))))))(((((((((((..(((...)))..))))))...((((.......))))))))).............................. (-28.77 = -29.13 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:31 2011