
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,023,772 – 18,023,864 |
| Length | 92 |
| Max. P | 0.562331 |

| Location | 18,023,772 – 18,023,864 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 64.47 |
| Shannon entropy | 0.63142 |
| G+C content | 0.53259 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -11.39 |
| Energy contribution | -11.61 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18023772 92 - 27905053 AAAUGCAAGUCAGGCGCCGA-UGGCACGCAAGUGAUCAUCACACCUUGAUAAAGAGGA-GGAGUU------------------GGAGGACUACAGGGCUACCCCAUCUCAAU .......((((.(((.((((-((((((....))).)))))...((((......)))).-)).)))------------------....))))...(((....)))........ ( -28.10, z-score = -1.52, R) >droSim1.chr3R 17840131 98 - 27517382 AAAUGCAAGUCAGGCGCCGA-UGGCACGCAAGUGAUCAUCACACCUUGAUAAAGAGGA-GGAGUUGGAGUU------------GGAGGACUACAGGGCUACCCCAUCUCAAU .....(((.((..((.((((-((((((....))).)))))...((((......)))).-)).))..)).))------------)((((......(((....))).))))... ( -29.90, z-score = -1.22, R) >droSec1.super_0 18393238 98 - 21120651 AAAUGCAAGUCAGGCGCCGA-UGGCACGCAAGUGAUCAUCACACCUUGAUAAAGAGGA-GGAGUUGGAGUU------------GGAGGACUACAAGGCUACCCCAUCUCAAU ........((((((....((-((((((....))).)))))....))))))...(((..-((.(.(((..((------------(........)))..))).)))..)))... ( -28.00, z-score = -0.95, R) >droYak2.chr3R 18874205 110 - 28832112 AAAUGCAAGUCAGGCGCCGA-UGGCACGCAAGUGAUCAUCACACCUUGAUAAAGAGGA-GGAGUUGGAGUUGAAGUUGGAGUUGGAGGACUACAGGGCUACCCCAUCUCAAU .....(((.((..((.((((-((((((....))).)))))...((((......)))).-)).))..)).)))......(((.(((.((............))))).)))... ( -33.50, z-score = -1.14, R) >droAna3.scaffold_13340 9021699 90 - 23697760 AAAUGCAAGUCAGGCGCCGA-UGGCACGCAAGUGAUCAUCACACCUUGAUAAAGAUGAGGAGGCAGGGGCUUUGAUUCCCCCCAUCUCAAU--------------------- ........((((((....((-((((((....))).)))))....)))))).....((((..((..((((........))))))..))))..--------------------- ( -29.50, z-score = -1.48, R) >dp4.chr2 12210448 71 - 30794189 AAAUGCCAGUCAGGCGCUGAGUGGCACGCAAGUGAUCAUCACACCUUGAUAAGGAGAG-GGUACCCCCCGCC---------------------------------------- ...(((((.((((...)))).))))).((..(((.....))).(((.....)))...(-((.....))))).---------------------------------------- ( -22.30, z-score = -1.10, R) >droPer1.super_0 3534795 71 + 11822988 AAAUGCAAUCCAGGCGCCGAGUGGCACGCAAGUGUUCAUCACACCUUGAUAAGAGAAG-GGUACCCCCCCCC---------------------------------------- .......(((.(((.(..((..(((((....)))))..)).).))).))).......(-((......)))..---------------------------------------- ( -17.30, z-score = -0.64, R) >droVir3.scaffold_13047 10771855 70 + 19223366 AAAUGCAAGUCAGGCGCCGC-UGGCACGCAAGUGAUCAUCACACCUUGAUAAGGUACU-GGGACUCGAGAGC---------------------------------------- .....(.((((.((..((..-...(((....)))((((........))))..))..))-..)))).).....---------------------------------------- ( -17.20, z-score = 0.40, R) >droMoj3.scaffold_6540 27716318 71 - 34148556 AAAUGCAAGUCAGGCGCCGC-UGGCACGCAAGUGAUCAUCACACCUUGAUAAGGUGCUCGGGACGCGAGAGC---------------------------------------- ........(((.((((((..-...(((....)))((((........))))..))))))...)))((....))---------------------------------------- ( -23.40, z-score = -0.78, R) >consensus AAAUGCAAGUCAGGCGCCGA_UGGCACGCAAGUGAUCAUCACACCUUGAUAAAGAGGA_GGAGCUCGAGUUC________________A_U_____________________ ........((((((.(.....((((((....))).))).....))))))).............................................................. (-11.39 = -11.61 + 0.22)

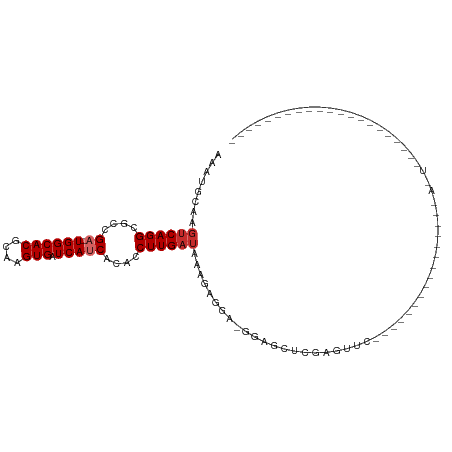
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:26 2011