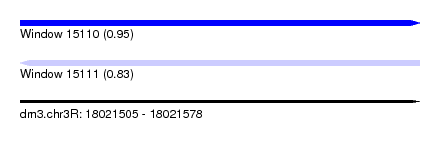
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 18,021,505 – 18,021,578 |
| Length | 73 |
| Max. P | 0.946527 |

| Location | 18,021,505 – 18,021,578 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 66.40 |
| Shannon entropy | 0.66545 |
| G+C content | 0.49691 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -9.61 |
| Energy contribution | -10.34 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
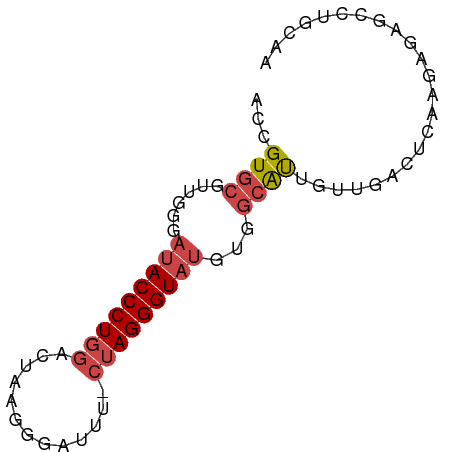
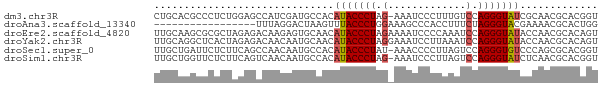
>dm3.chr3R 18021505 73 + 27905053 ACCGUGCGUUGCGAUACCCUGGACAAAGGGAUUU-CUAGGGUAUGUGGCAUCGAUGGCUCCAGAGGGCGUGCAG ...(..(((..(.((((((((((..........)-))))))))))..)).......((((....)))))..).. ( -27.60, z-score = -1.62, R) >droAna3.scaffold_13340 9019725 57 + 23697760 CCAGUGCGUUUUCGUACCCUAGAAAGGUGGGCUUUCCAGGGUAAACUUAGUCCUAAA----------------- ..((..(.......((((((.(((((.....))))).))))))......)..))...----------------- ( -15.92, z-score = -1.24, R) >droEre2.scaffold_4820 414373 74 - 10470090 ACUGUGCGUUGGUAUACCCUGGAUUUGGGGAUUUUCUAGGGUAUGUUGCACUCUUGUCUCUAGCGCGCUUGCAA ...((((((((((((((((((((...........)))))))))))..(((....)))..)))))))))...... ( -26.80, z-score = -3.07, R) >droYak2.chr3R 18871901 74 + 28832112 ACUGUGCGUUGGUAUACCCUGGAUUUAAGGAUUUCCUAGGGUAUGUUGCAUUGUUGUCUCUAGUGAGCCUGCAA ...(((((....(((((((((((..........))).)))))))).)))))(((.(.(((....))).).))). ( -21.50, z-score = -1.55, R) >droSec1.super_0 18391031 73 + 21120651 ACCGUGCGCUGGGACACCCUGGACUAAGGGGUUU-AUAGGGUAUGUGGCAUUGUUGGCUGAAGAGAAUCAGCAA .((((((.(((((((.((((......))))))))-.))).))))).).........(((((......))))).. ( -23.90, z-score = -1.29, R) >droSim1.chr3R 17837901 73 + 27517382 ACCGUGCGUUGAGAUACCCUGGACUAAGGGAUUU-CUAGGGUAUGUGGCAUUGUUGACUGAAGAGAACCAGCAA .((((((.(((((((.((((......))))))))-).)).))))).)...((((((............)))))) ( -19.00, z-score = -0.75, R) >consensus ACCGUGCGUUGGGAUACCCUGGACUAAGGGAUUU_CUAGGGUAUGUGGCAUUGUUGACUCAAGAGAGCCUGCAA ...((((......(((((((((.............)))))))))...))))....................... ( -9.61 = -10.34 + 0.72)

| Location | 18,021,505 – 18,021,578 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 66.40 |
| Shannon entropy | 0.66545 |
| G+C content | 0.49691 |
| Mean single sequence MFE | -16.50 |
| Consensus MFE | -6.34 |
| Energy contribution | -6.36 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 18021505 73 - 27905053 CUGCACGCCCUCUGGAGCCAUCGAUGCCACAUACCCUAG-AAAUCCCUUUGUCCAGGGUAUCGCAACGCACGGU .(((..((((...)).))....(.(((...(((((((.(-(.(......).)).))))))).))).)))).... ( -16.20, z-score = 0.41, R) >droAna3.scaffold_13340 9019725 57 - 23697760 -----------------UUUAGGACUAAGUUUACCCUGGAAAGCCCACCUUUCUAGGGUACGAAAACGCACUGG -----------------..(((..(....((((((((((((((.....)))))))))))).))....)..))). ( -17.30, z-score = -2.13, R) >droEre2.scaffold_4820 414373 74 + 10470090 UUGCAAGCGCGCUAGAGACAAGAGUGCAACAUACCCUAGAAAAUCCCCAAAUCCAGGGUAUACCAACGCACAGU .(((..((((.((.......)).))))...(((((((.((...........)).)))))))......))).... ( -17.90, z-score = -3.08, R) >droYak2.chr3R 18871901 74 - 28832112 UUGCAGGCUCACUAGAGACAACAAUGCAACAUACCCUAGGAAAUCCUUAAAUCCAGGGUAUACCAACGCACAGU (((((.((((....))).).....))))).(((((((.(((..........))))))))))............. ( -17.60, z-score = -2.57, R) >droSec1.super_0 18391031 73 - 21120651 UUGCUGAUUCUCUUCAGCCAACAAUGCCACAUACCCUAU-AAACCCCUUAGUCCAGGGUGUCCCAGCGCACGGU ..(((((......)))))..((..((((..(((((((.(-((.....)))....)))))))....).)))..)) ( -15.80, z-score = -1.02, R) >droSim1.chr3R 17837901 73 - 27517382 UUGCUGGUUCUCUUCAGUCAACAAUGCCACAUACCCUAG-AAAUCCCUUAGUCCAGGGUAUCUCAACGCACGGU ..(((((......)))))..((..(((...(((((((.(-(..........)).)))))))......)))..)) ( -14.20, z-score = -0.67, R) >consensus UUGCAGGCUCUCUAGAGACAACAAUGCAACAUACCCUAG_AAAUCCCUUAGUCCAGGGUAUCCCAACGCACGGU ..............................(((((((.................)))))))............. ( -6.34 = -6.36 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:26 2011