| Sequence ID | dm3.chr3R |
|---|---|
| Location | 17,981,790 – 17,981,881 |
| Length | 91 |
| Max. P | 0.632767 |

| Location | 17,981,790 – 17,981,881 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 64.91 |
| Shannon entropy | 0.71897 |
| G+C content | 0.32115 |
| Mean single sequence MFE | -19.09 |
| Consensus MFE | -5.81 |
| Energy contribution | -5.73 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632767 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
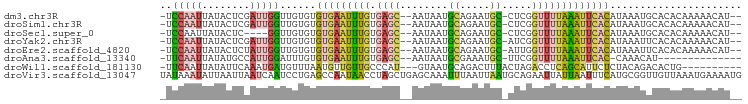
>dm3.chr3R 17981790 91 - 27905053 -UCCAAUUAUACUCGAUUGGUUGUGUGUGAAUUUGUGAGC--AAUAAUGCAGAAUGC-CUCGGUUUUAAAUUCACAUAAAUGCACACAAAAACAU-- -.((((((......))))))((((((((..(((((((.((--(....))).((((((-....)).....)))).)))))))))))))))......-- ( -22.70, z-score = -2.15, R) >droSim1.chr3R 17798234 91 - 27517382 -UCCAAUUAUACUCGAUUGGUUGUGUGUGAAUUUGUGAGC--AAUAAUGCAGAAUGC-CUCGGUUUUAAAUUCACAUAAAUGCACACAAAAACAU-- -.((((((......))))))((((((((..(((((((.((--(....))).((((((-....)).....)))).)))))))))))))))......-- ( -22.70, z-score = -2.15, R) >droSec1.super_0 18351758 87 - 21120651 -UCCAAUUAUACUC----GGUUGUGUGUGAAUUUGUGAGC--AAUAAUGCAGAAUGC-CUCGGUUUUAAAUUCACAUAAAUGCACACAAAAACAU-- -.............----.(((.((((((.(((((((.((--(....))).((((((-....)).....)))).))))))).))))))..)))..-- ( -18.80, z-score = -1.18, R) >droYak2.chr3R 18830926 91 - 28832112 -UCCAAUUAUACUCGAUUGGUUGUGUGUGAAUUUGUGAGC--AAUAAUGCAGAAUGC-AUCGGUUUUAAAUUCACAUAAAUUCACACAAAAACAU-- -.((((((......))))))...((((((((((((((.((--(....))).((((..-...........)))).)))))))))))))).......-- ( -26.22, z-score = -3.69, R) >droEre2.scaffold_4820 374289 91 + 10470090 -UCCAAUUAUACUCUAUUGGUUGUGUGUGAAUUUGUGAGC--AAUAAUGCAGAAUGC-AUUGGUUUUAAAUUCACAUAAAUUCACACAAAAACAU-- -.(((((........)))))...((((((((((((((.((--(....))).((((..-...........)))).)))))))))))))).......-- ( -25.22, z-score = -3.43, R) >droAna3.scaffold_13340 8981791 78 - 23697760 -UUCAAUUAUAUGCCAUUGGAUUUGUGUGAAUUUGUGAGC--AAUAAUGCGAAAUGC-UUCGGUUUUAAAUUCAC-CAAACAU-------------- -((((((........))))))((((.(((((((((...((--(....))).....((-....))..)))))))))-))))...-------------- ( -16.00, z-score = -0.88, R) >droWil1.scaffold_181130 12122941 83 - 16660200 -UUCAAUUAUAUUCAAAUGAUGUUUAAUGUUGUUGCCCAU---GUAAUGCAGACUUUACUAGACCUCAGCAUUCUCUACAGACACUG---------- -...................(((..(((((((........---((((........)))).......)))))))....))).......---------- ( -8.36, z-score = 1.23, R) >droVir3.scaffold_13047 10725904 97 + 19223366 UAUAAAUAUUAAUUAAUCAAUCCUGAGCCAAUAACCUAGCUGAGCAAAUUUAAUUAAUGCAGAAUUAUUAAUUUCAUGCGGUUGUUAAAUGAAAAUG ................((....((.(((..........))).))...(((((((...(((((((........))).))))...)))))))))..... ( -12.70, z-score = -0.05, R) >consensus _UCCAAUUAUACUCGAUUGGUUGUGUGUGAAUUUGUGAGC__AAUAAUGCAGAAUGC_CUCGGUUUUAAAUUCACAUAAAUGCACACAAAAACAU__ ..(((((........)))))......(((((((((.((((........((.....)).....)))))))))))))...................... ( -5.81 = -5.73 + -0.08)

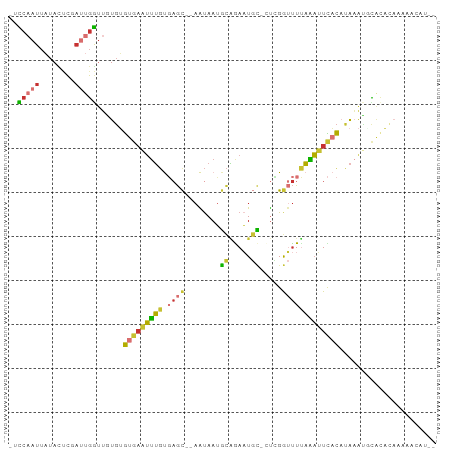
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:34:22 2011